Structural motifs in the PDB
Question: Has anyone see a disulfide forming between the two cys of "CXC" in the middle of a loop, and create a sharp turn, where X is not a proline? I seems to me that geometrically this would be possible but I am not sure how stable it is, or how energetically unfavorable it might be.
This is the kind of task that can be carried out quickly and easily with PDBeMotif - and here's how to do it:
- Surf to PDBeMotif at http://pdbe.org/motif
- Click on the "Search" tab (or the "Search" link)
- Select the option to search "Protein sequence, pattern" by clicking on the corresponding icon on the left of the page (fifth icon down)
- In the sequence box enter: CXC
- Select the radio button to search using "Pattern match on observed PDB sequences"
- Click on "Add it to the search criteria"
If we were to carry out the current search, we would get all observed instances of Cys-X-Cys in the PDB. However, we want to specify that the two cysteines are covalently linked. This can be done as follows:
- Select the option to search "Interactions" by clicking on the corresponding icon on the left of the page (sixth icon down)
- In the panel, change the two drop-down menus labelled "Interacting residue" so that one says "1" and the other says "3" (from their default value "any"). Also set the "Bond type" drop-down to "Covalent bond".
- Click on "Add it to the search criteria"
Now we have specified that we want to find instances of Cys-X-Cys, where there is a covalent bond between the two Cys residues (residues 1 and 3 in the pattern CXC).
- Before you hit the "Search" button, you can limit the search with the "Normalise hits by" drop-down. If you select "-" you will get all instances in the PDB; if you select "UniProt", only one entry per UniProt ID will be retrieved, etc.
- Hit the "Search" button
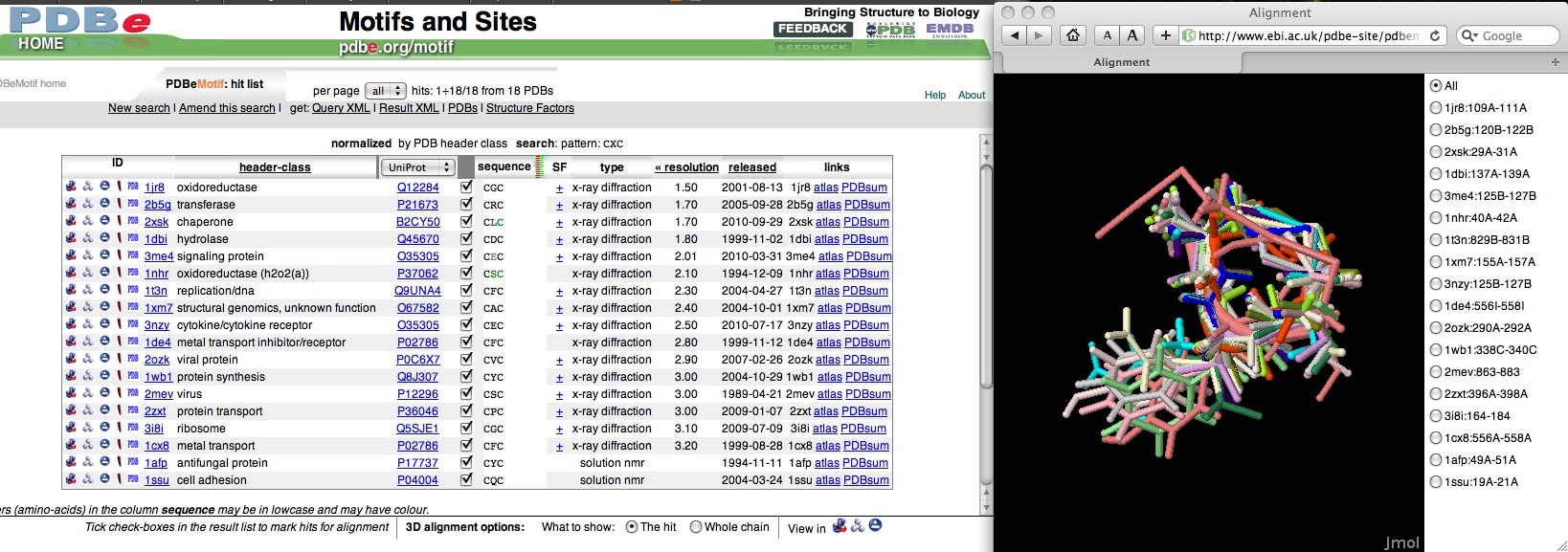
- If you searched for all instances in the PDB, PDBeMotif will return a list of 147 hits in 55 PDB entries. If you normalised by PDB header, you will get 18 hits in 18 entries.
- To visualise (some of) the hits, mark the check boxes of the instances you're interested in. Then select a visualisation program at the bottom of the page by clicking on its icon (e.g., Jmol). All the selected hits will be superimposed and shown in the selected viewer.
A screendump (you'll have to scroll to the right to see the superposition!):
(Note: if you're an advanced user, you may one day want to edit and submit your queries in XML - click on "Get Query XML" for more information.)