Solve a small-molecule structure: Difference between revisions
No edit summary |
|||
| Line 163: | Line 163: | ||
ANIS | ANIS | ||
line into 56.ins . More info about refinement options is in the [[SHELXL]] article! | line into 56.ins . More info about refinement options is in the [[SHELXL]] article! | ||
== Electron density == | |||
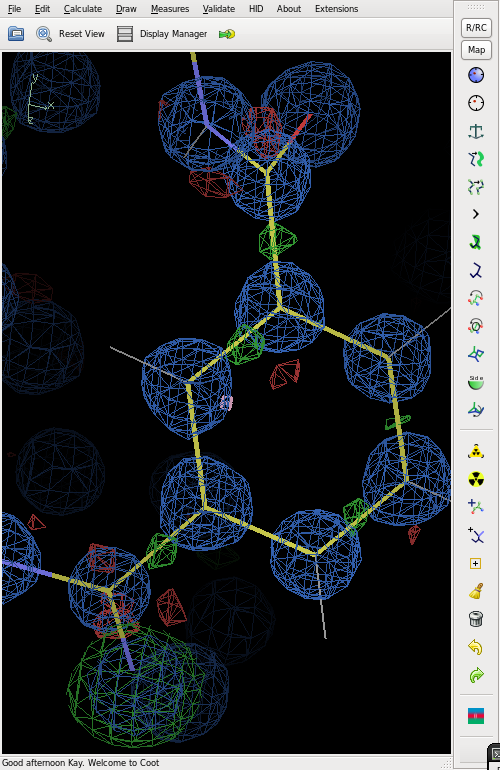
The figure shows the final electron density (blue), but with an O atom refined as N. This gives strong positive (green) difference electron density. | |||
[[File:Diffden.png]] | |||
The difference map also shows distinct bonding electron density on most of the bonds. | |||
Revision as of 12:08, 1 March 2011
The following is based on the experience of a protein crystallographer who one day obtained a small-molecule dataset and managed to solve and refine it without prior knowledge what the crystallized substance was. It was a very rewarding experience which is why it's written up here.
This is just a case study. To understand things, one has to read http://shelx.uni-ac.gwdg.de/SHELX/shelx.pdf .
reduce the data with your favourite data processing software
I use XDS. The decision about the spacegroup has to be postponed, but it surely helps if the correct Laue group is employed during scaling. In the case considered here, the CORRECT step suggested P222.
convert the reflection file to HKLF 4 format (intensities!)
The HKLF 4 format is what the SHELX programs read. I used XDSCONV and the following XDSCONV.INP:
INPUT_FILE=XDS_ASCII.HKL OUTPUT_FILE=temp.hkl
first try: wrong spacegroup
run XPREP to find out possible spacegroups
answer the question concerning the cell axes, and then hit <Enter> several times until the program suggests a list of spacegroups - this choice is going to be important. It helps a bit to observe earlier whether it's centrosymmetric or not, from the line: Mean |E*E-1| = 0.939 [expected .968 centrosym and .736 non-centrosym].
Systematic absence exceptions:
b-- c-- n-- 21-- -c- -a- -n- -21- --a --b --n --21
N 938 938 0 0 411 0 411 0 0 237 237 0
N I>3s 706 706 0 0 304 0 304 0 0 203 203 0
<I> 50.0 50.0 0.0 0.0 43.1 0.0 43.1 0.0 0.0 56.6 56.6 0.0
<I/s> 14.1 14.1 0.0 0.0 15.2 0.0 15.2 0.0 0.0 16.4 16.4 0.0
Identical indices and Friedel opposites combined before calculating R(sym)
Option Space Group No. Type Axes CSD R(sym) N(eq) Syst. Abs. CFOM
[A] P222 # 16 chiral 1 14 0.022 9725 0.0 / 10.7 11.72
[B] Pmm2 # 25 non-cen 1 9 0.022 9725 0.0 / 10.7 15.05
[C] Pmm2 # 25 non-cen 5 9 0.022 9725 0.0 / 10.7 15.05
[D] Pmm2 # 25 non-cen 3 9 0.022 9725 0.0 / 10.7 15.05
[E] Pmmm # 47 centro 1 7 0.022 9725 0.0 / 10.7 13.52
[F] P222(1) # 17 chiral 1 26 0.022 9725 0.0 / 10.7 8.76
[G] P222(1) # 17 chiral 5 26 0.022 9725 0.0 / 10.7 8.76
[H] P222(1) # 17 chiral 3 26 0.022 9725 0.0 / 10.7 8.76
[I] P2(1)2(1)2 # 18 chiral 1 359 0.022 9725 0.0 / 10.7 5.33
[J] P2(1)2(1)2 # 18 chiral 5 359 0.022 9725 0.0 / 10.7 5.33
[K] P2(1)2(1)2 # 18 chiral 3 359 0.022 9725 0.0 / 10.7 5.33
[L] P2(1)2(1)2(1) # 19 chiral 1 5917 0.022 9725 0.0 / 10.7 5.07
[M] Pmc2(1) # 26 non-cen 3 20 0.022 9725 0.0 / 10.7 9.81
[N] Pmc2(1) # 26 non-cen 4 20 0.022 9725 0.0 / 10.7 9.81
[O] Pmma # 51 centro 1 14 0.022 9725 0.0 / 10.7 7.69
[P] Pmma # 51 centro 6 14 0.022 9725 0.0 / 10.7 7.69
[R] Pma2 # 28 non-cen 1 1 0.022 9725 0.0 / 10.7 55.05
[S] Pma2 # 28 non-cen 6 1 0.022 9725 0.0 / 10.7 55.05
[T] Pmn2(1) # 31 non-cen 2 53 0.022 9725 0.0 / 10.7 6.90
[U] Pmn2(1) # 31 non-cen 5 53 0.022 9725 0.0 / 10.7 6.90
[V] Pmmn # 59 centro 3 42 0.022 9725 0.0 / 10.7 3.35
[W] Pcc2 # 27 non-cen 3 2 0.022 9725 0.0 / 10.7 38.39
[X] Pccm # 49 centro 3 1 0.022 9725 0.0 / 10.7 51.02
[Y] Pna2(1) # 33 non-cen 1 903 0.022 9725 0.0 / 10.7 5.16
[Z] Pna2(1) # 33 non-cen 6 903 0.022 9725 0.0 / 10.7 5.16
[0] Pnma # 62 centro 1 894 0.022 9725 0.0 / 10.7 1.14
[1] Pnma # 62 centro 6 894 0.022 9725 0.0 / 10.7 1.14
[2] Pccn # 56 centro 3 196 0.022 9725 0.0 / 10.7 1.53
Option [1] chosen
(The program chooses Option "1" (Pnma) by default, which later turns out to be wrong. How the correct spacegroup (Pccn) could be identified at this point, I don't know.)
After that, say "c" for "define unit-cell CONTENTS", and input a reasonable number of carbon atoms (I used C20). Get out of this menu with "E". Then, choose "f" for "set up shelxtl FILES". Then, answer the question "XM/SHELXD (M) or XS/SHELXS (S) format [S]:" with "m" since we're going to use shelxd for solving the structure. Answer the question about the name (I used the spacegroup number as I knew I would have to test several possibilities). Finally, "q"uit the program. The resulting 62.ins is:
TITL 62 in Pnma CELL 0.71073 8.4900 28.7000 14.4300 90.000 90.000 90.000 ZERR 11.00 0.0017 0.0057 0.0029 0.000 0.000 0.000 LATT 1 SYMM 0.5-X, -Y, 0.5+Z SYMM -X, 0.5+Y, -Z SYMM 0.5+X, 0.5-Y, 0.5-Z SFAC C UNIT 220 FIND 16 PLOP 22 27 31 MIND 1.0 -0.1 NTRY 1000 HKLF 4 END
solving the structure with SHELXD
Just run "shelxd 62". You may interrupt it with Ctrl-C once it has found good solutions, as suggested by
Try 68:20 Peaks 99 96 71 68 63 55 53 51 50 48 46 45 45 44 44 43 43 43 41 40 R = 0.417, Min.fun. = 0.853, <cos> = 0.364, Ra = 0.432 Try 68, CC All/Weak 40.17 / 25.34, best 40.17 / 25.60, best final CC 0.00 Peaklist optimization cycle 1 CC = 46.01 % BG = 0.638 for 21 atoms Peaks: 99 91 66 63 63 55 54 49 49 45 43 43 42 42 41 39 20 18 18 17 17 -17 Fragments: 7 4 4 3 1 1 1 Peaklist optimization cycle 2 CC = 52.55 % BG = 0.593 for 25 atoms Peaks: 99 94 85 74 73 73 70 70 66 64 64 63 62 62 61 60 60 60 59 59 57 24 -24 Fragments: 14 5 4 1 1 Peaklist optimization cycle 3 CC = 58.37 % BG = 0.541 for 29 atoms Peaks: 99 92 85 72 72 70 69 66 65 63 63 62 61 60 59 59 59 -58 58 58 57 57 56 Fragments: 17 7 4 1
and the resulting 62.res is:
REM TRY 77 FINAL CC 58.70 TIME 5 SECS REM Fragments: 17 7 3 2 2 REM TITL 62 in Pnma CELL 0.71073 8.4900 28.7000 14.4300 90.000 90.000 90.000 ZERR 11.00 0.0017 0.0057 0.0029 0.000 0.000 0.000 LATT 1 SYMM 0.5-X, -Y, 0.5+Z SYMM -X, 0.5+Y, -Z SYMM 0.5+X, 0.5-Y, 0.5-Z SFAC C UNIT 220 C001 1 0.15479 0.75000 0.04294 10.50000 0.1 99.00 C002 1 0.84807 0.75000 -0.04054 10.50000 0.1 77.20 C003 1 0.19291 0.85742 -0.17716 11.00000 0.1 63.76 C004 1 0.59349 0.82735 0.20939 11.00000 0.1 62.64 C005 1 0.84406 0.88664 0.13204 11.00000 0.1 61.71 C006 1 0.23705 0.75000 -0.14287 10.50000 0.1 61.63 ... C026 1 0.72766 0.95461 -0.10200 11.00000 0.1 51.40 C027 1 0.77380 0.96500 0.10122 11.00000 0.1 49.11 C028 1 0.39642 0.72972 0.17524 11.00000 0.1 24.76 C029 1 0.66918 0.82482 0.13969 11.00000 0.1 21.87 C030 1 0.28518 0.73520 0.01792 11.00000 0.1 21.39 C031 1 0.40533 0.78770 0.08494 11.00000 0.1 19.94 HKLF 4 END
Refinement in SHELXL
In this example, it makes sense to remove the last 4 atoms since their occupancy is less than 25% of the maximum, and the final remaining atom then is at 49% - a large jump. Insert
ACTA LIST 6 L.S. 10
after the UNIT 220 instruction, and run "shelxl 62".
It turns out that the R-factor does not really go down properly, and this means that the spacegroup is wrong. The "FINAL CC 58.70" result from SHELXD is probably also suspiciously low, I guess.
structure solution and refinement in the correct spacegroup
We have to go back to XPREP and try a different spacegroup. This time I use Option "2" which means "Pccn" (number 56). SHELXD (finding 25 atoms, with a "FINAL CC 88.86") and SHELXL are run in the same way as above, but this time the R1 goes down to something above 10%, which indicates that this is probably a solution.
general idea of refining a structure
Starting from a rough guess of the number of atoms, we adjust the model, guided by the refinement results. This is an iterative process, in which we repeatedly edit 56.res to reflect our change of conception of the structure, replace 56.ins with it, and run SHELXL again.
assigning chemical types
Since we know that there's not only carbon atoms, but likely also N, O and H, we modify 56.ins to have
SFAC C N O H UNIT 200 100 100 40
(the actual numbers after UNIT can be taken from the .lst file of SHELXL, they don't seem to matter much.)
We tell SHELXL the chemical identity by putting a 1 for a C, a 2 for a N, a 3 for an O, and a 4 for a H - the number is just the order of the atom in the SFAC line.
The chemical identity of an atom can be found from geometric parameters, and its electron density. The electron density can be displayed e.g. in coot, by loading the 56.fcf file written by SHELXL. Geometric parameters (in particular distances) are listed in the 56.lst file. Typical bond distances of C-C, C=C, C-O, C=O, C-N, X-H are .....
As a proxy to electron density we can use the refined ADPs. Atoms initially called "C", but with very low U values after refinement, are most likely O or N atoms.
For the H atoms, we just cut-and-paste the atoms from the bottom of the .res file into those lines where the other atoms are, if the distances to existing (heavy) atoms are close to 1 A.
Finishing the structure
Finally we switch to anisotropic refinement by putting an
ANIS
line into 56.ins . More info about refinement options is in the SHELXL article!
Electron density
The figure shows the final electron density (blue), but with an O atom refined as N. This gives strong positive (green) difference electron density.
The difference map also shows distinct bonding electron density on most of the bonds.