Solve a small-molecule structure: Difference between revisions
(→Solve the structure with SHELXD: add GS posting) |
Lazarenko VA (talk | contribs) |
||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
Maybe it should also be stated that this was a simple case, without e.g. twinning or disorder! Furthermore, the hand of the structure was not an issue. | Maybe it should also be stated that this was a simple case, without e.g. twinning or disorder! Furthermore, the hand of the structure was not an issue. | ||
== Collecting data == | |||
There's not much magic in collecting data. Problems arise from the high resolution that's required and the strength and low number of reflections. | |||
* Mount the crystal by gluing it on a steel pin. More refined approaches might exist. | |||
* High resolution is important. 0.84Å is the minimum for publication in Acta Cryst. 1.2Å is the absolute minimum for structure solution. This can generally only be achieved on a PX system with a detector on a two-theta arm. | |||
* Crystal quality is important. There should be no streaky spots, multiple lattices, etc. You can always break off small pieces if the big chunk isn't clean enough. | |||
* As there are only few spots per image, a large rotation range is usually needed for indexing. Collect ten degrees in one-degree oscillations. This is better than collecting one ten-degree oscillation because the phi angle of each reflection is more accurately determined and the background is lower. | |||
* The beam might need to be attenuated to avoid overloads. This can be done by dialing down the energy of the electron beam going into the anode. | |||
* If a heavy atom (iodine, iron, etc.) is present in the small molecule, the data can probably be phased by SAD even with Cu Kalpha. You might be able to solve it by looking at the Patterson maps. | |||
== Reduce the data with your favourite data processing software == | == Reduce the data with your favourite data processing software == | ||
| Line 19: | Line 30: | ||
=== use [[XPREP]] to find possible spacegroups === | === use [[XPREP]] to find possible spacegroups === | ||
There is no longer a need to use [[xds:XDSCONV|XDSCONV]] to convert the XDS_ASCII.HKL reflection file to HKLF 4 format (which is what the SHELX programs read) since XPREP can read XDS_ASCII.HKL directly. Just run | |||
xprep | |||
without a filename, and when the filename prompt appears, enter: | |||
XDS_ASCII.HKL | |||
(or whatever you have renamed it to) and then hit <Enter> several times until the program suggests a list of spacegroups - this list is going to be important. It may help to observe whether the data are centrosymmetric or not, from the 8th non-blank line below. Fortunately, this time there's only one spacegroup consistent with the data: | |||
<pre> | <pre> | ||
SPACE GROUP DETERMINATION | SPACE GROUP DETERMINATION | ||
| Line 66: | Line 74: | ||
</pre> | </pre> | ||
After that, say "c" for "define unit-cell CONTENTS", and input a reasonable number of carbon atoms (I used C20). Get out of this menu with "E". Then, choose "f" for "set up shelxtl FILES". Then, answer the question "XM/SHELXD (M) or XS/SHELXS (S) format [S]:" with "m" since we're going to use shelxd for solving the structure. Answer the question about the name (I used the spacegroup number as I knew I would have to test several possibilities). Finally, "q"uit the program. This writes 56.ins : | After that, say "c" for "define unit-cell CONTENTS", and input a reasonable number of carbon atoms (I used C20). After that you will probably need to change the wavelength, because by default xprep use Mo K_alpha, you can do it by saying "R". Get out of this menu with "E". Then, choose "f" for "set up shelxtl FILES". Then, answer the question "XM/SHELXD (M) or XS/SHELXS (S) format [S]:" with "m" since we're going to use shelxd for solving the structure. Answer the question about the name (I used the spacegroup number as I knew I would have to test several possibilities). Finally, "q"uit the program. This writes 56.ins : | ||
TITL 56 in Pccn | TITL 56 in Pccn | ||
CELL 0.71073 14.4330 28.7040 8.4880 90.000 90.000 90.000 | CELL 0.71073 14.4330 28.7040 8.4880 90.000 90.000 90.000 | ||
| Line 82: | Line 90: | ||
HKLF 4 | HKLF 4 | ||
END | END | ||
Compared to the P1 setting that CORRECT chose, XPREP has re-indexed the data in this example such that the conventional setting is obtained for this space group. | |||
If necessary XPREP can read in several XDS_ASCII.HKL files, scale them together and merge them. However it needs to start with one file to get the space group so that it knows how to merge. | |||
=== use [[ccp4dev:Symmetry_determination_with_Pointless|POINTLESS]] to find possible spacegroups === | === use [[ccp4dev:Symmetry_determination_with_Pointless|POINTLESS]] to find possible spacegroups === | ||
| Line 139: | Line 151: | ||
UNIT_CELL_CONSTANTS= 14.433 28.704 8.488 90.000 90.000 90.000 | UNIT_CELL_CONSTANTS= 14.433 28.704 8.488 90.000 90.000 90.000 | ||
INPUT_FILE=XDS_ASCII.HKL | INPUT_FILE=XDS_ASCII.HKL | ||
OUTPUT_FILE=56.hkl | OUTPUT_FILE=56.hkl SHELX | ||
Please note that the file 56.ins has to be set up manually in this case (just take the 56.ins from above, and adjust the symops and cell parameters). The numbers after "FIND" and "PLOP" should probably be adjusted in proportion to the expected number of atoms in the asymmetric unit. | Please note that the file 56.ins has to be set up manually in this case (just take the 56.ins from above, and adjust the symops and cell parameters). The numbers after "FIND" and "PLOP" should probably be adjusted in proportion to the expected number of atoms in the asymmetric unit. | ||
| Line 231: | Line 243: | ||
As a proxy to electron density we can use the refined ADPs. Atoms initially called "C", but with very low U values after refinement, are most likely O or N atoms. | As a proxy to electron density we can use the refined ADPs. Atoms initially called "C", but with very low U values after refinement, are most likely O or N atoms. | ||
For the H atoms, we just | |||
=== Hydrogens === | |||
For the H atoms, we just move the atoms from the bottom of the .res file to those lines where the refined atoms are, if the distances to existing (heavy) atoms are close to 1 A. For hydrogens bond to C N O (and in some cases B) we could alternatively use the HFIX instruction which sets up suitable AFIX instructions for the standard 'riding H-atom' refinement (shelXle - see below - can do this with one click). This requires lines of the form | |||
HFIX 13 XXX | |||
In this example; the first digit 1 means tert-CH (2 would mean methylen-CH2, 3 would mean methyl-CH3, 4 would mean aromatic CH), and the second digit 3 means the normal riding model. XXX stands for the (heavy) atom name. For docs and more examples see [https://www.google.com/search?btnG=1&pws=0&q=hfix+shelxl]. | |||
=== Finishing the structure === | === Finishing the structure === | ||
| Line 257: | Line 274: | ||
The difference map also shows distinct bonding electron density on most of the bonds. | The difference map also shows distinct bonding electron density on most of the bonds. | ||
== A GUI for refining small molecule structures == | |||
A GUI called shelXle written by Christian Huebschle is now available for refining small molecule crystal structures with shelxl: http://ewald.ac.chemie.uni-goettingen.de/shelx | |||
Latest revision as of 10:11, 19 October 2020
The following is based on the experience of a protein crystallographer who one day obtained a small-molecule dataset and managed to solve and refine it without prior knowledge what the crystallized substance was, and without experience in small-molecule crystallography. It was a very rewarding experience (see the figure at the bottom) which is why it's written up here.
This writeup is only meant for the protein crystallographer who occasionally has to use existing tools on a small-molecule dataset. To understand things more thoroughly, one has to read http://shelx.uni-ac.gwdg.de/SHELX/shelx.pdf . There are lots of tutorials available from George Sheldrick's website, but also from others, e.g. [1].
Maybe it should also be stated that this was a simple case, without e.g. twinning or disorder! Furthermore, the hand of the structure was not an issue.
Collecting data[edit | edit source]
There's not much magic in collecting data. Problems arise from the high resolution that's required and the strength and low number of reflections.
- Mount the crystal by gluing it on a steel pin. More refined approaches might exist.
- High resolution is important. 0.84Å is the minimum for publication in Acta Cryst. 1.2Å is the absolute minimum for structure solution. This can generally only be achieved on a PX system with a detector on a two-theta arm.
- Crystal quality is important. There should be no streaky spots, multiple lattices, etc. You can always break off small pieces if the big chunk isn't clean enough.
- As there are only few spots per image, a large rotation range is usually needed for indexing. Collect ten degrees in one-degree oscillations. This is better than collecting one ten-degree oscillation because the phi angle of each reflection is more accurately determined and the background is lower.
- The beam might need to be attenuated to avoid overloads. This can be done by dialing down the energy of the electron beam going into the anode.
- If a heavy atom (iodine, iron, etc.) is present in the small molecule, the data can probably be phased by SAD even with Cu Kalpha. You might be able to solve it by looking at the Patterson maps.
Reduce the data with your favourite data processing software[edit | edit source]
I use XDS. The decision about the spacegroup has to be postponed, but it surely helps if the correct Laue group is employed during scaling. In the case considered here, the CORRECT step suggested P222 (XDS really only should suggest "222 point symmetry" because CORRECT does not look at systematic absences at this point).
Determine the spacegroup[edit | edit source]
There are two ways to determine the spacegroup:
These two possibilities also differ in the way how to obtain a file suitable for input to the SHELX program.
If there are different spacegroup possibilities then (downstream, in structure solution and refinement) we need to try all of them in turn, until we hit one that refines really satisfactorily (R-factor below, say, 5%) and gives a structure that makes sense.
use XPREP to find possible spacegroups[edit | edit source]
There is no longer a need to use XDSCONV to convert the XDS_ASCII.HKL reflection file to HKLF 4 format (which is what the SHELX programs read) since XPREP can read XDS_ASCII.HKL directly. Just run
xprep
without a filename, and when the filename prompt appears, enter:
XDS_ASCII.HKL
(or whatever you have renamed it to) and then hit <Enter> several times until the program suggests a list of spacegroups - this list is going to be important. It may help to observe whether the data are centrosymmetric or not, from the 8th non-blank line below. Fortunately, this time there's only one spacegroup consistent with the data:
SPACE GROUP DETERMINATION
Lattice exceptions: P A B C I F Obv Rev All
N (total) = 0 28832 28824 28788 28823 43222 38376 38344 57564
N (int>3sigma) = 0 17961 18421 18158 17862 27270 24715 24627 36959
Mean intensity = 0.0 22.7 23.7 24.8 23.4 23.7 24.7 24.8 24.8
Mean int/sigma = 0.0 9.6 10.0 9.9 9.6 9.8 10.0 10.0 10.0
Crystal system O and Lattice type P selected
Mean |E*E-1| = 0.939 [expected .968 centrosym and .736 non-centrosym]
Chiral flag NOT set
Systematic absence exceptions:
b-- c-- n-- 21-- -c- -a- -n- -21- --a --b --n --21
N 1884 1884 1892 7 988 1014 992 28 545 541 534 72
N I>3s 706 706 0 0 304 0 304 0 0 203 203 0
<I> 25.2 25.2 0.5 0.0 18.2 0.4 18.1 0.4 0.4 25.0 25.4 0.4
<I/s> 7.3 7.3 0.5 0.2 6.6 0.5 6.6 0.5 0.4 7.4 7.6 0.4
Identical indices and Friedel opposites combined before calculating R(sym)
Option Space Group No. Type Axes CSD R(sym) N(eq) Syst. Abs. CFOM
[A] Pccn # 56 centro 3 196 0.023 10123 0.5 / 6.6 2.23
Option [A] chosen
After that, say "c" for "define unit-cell CONTENTS", and input a reasonable number of carbon atoms (I used C20). After that you will probably need to change the wavelength, because by default xprep use Mo K_alpha, you can do it by saying "R". Get out of this menu with "E". Then, choose "f" for "set up shelxtl FILES". Then, answer the question "XM/SHELXD (M) or XS/SHELXS (S) format [S]:" with "m" since we're going to use shelxd for solving the structure. Answer the question about the name (I used the spacegroup number as I knew I would have to test several possibilities). Finally, "q"uit the program. This writes 56.ins :
TITL 56 in Pccn CELL 0.71073 14.4330 28.7040 8.4880 90.000 90.000 90.000 ZERR 11.00 0.0029 0.0057 0.0017 0.000 0.000 0.000 LATT 1 SYMM 0.5-X, 0.5-Y, Z SYMM -X, 0.5+Y, 0.5-Z SYMM 0.5+X, -Y, 0.5-Z SFAC C UNIT 220 FIND 16 PLOP 22 27 31 MIND 1.0 -0.1 NTRY 1000 HKLF 4 END
Compared to the P1 setting that CORRECT chose, XPREP has re-indexed the data in this example such that the conventional setting is obtained for this space group.
If necessary XPREP can read in several XDS_ASCII.HKL files, scale them together and merge them. However it needs to start with one file to get the space group so that it knows how to merge.
use POINTLESS to find possible spacegroups[edit | edit source]
Unless the spacegroup number in XDS_ASCII.HKL already indicates this, pointless needs to be told that the spacegroup may not be restricted to those 65 which occur for crystals from macromolecules:
echo CHIRALITY NONCHIRAL | pointless xdsin XDS_ASCII.HKL
gives
Zone Number PeakHeight SD Probability ReflectionCondition
Zones for Laue group P m m m
1 screw axis 2(1) [a] 11 0.990 0.135 *** 0.972 h00: h=2n
2 screw axis 2(1) [b] 59 1.000 0.097 *** 0.986 0k0: k=2n
3 screw axis 2(1) [c] 131 0.997 0.062 *** 0.994 00l: l=2n
4 glide plane b(a) 3754 0.012 0.050 0.000 0kl: k=2n
5 glide plane c(a) 3754 0.013 0.050 0.000 0kl: l=2n
6 glide plane n(a) 3754 0.951 0.061 *** 0.988 0kl: k+l=2n
7 glide plane a(b) 1961 0.953 0.050 *** 0.990 h0l: h=2n
8 glide plane c(b) 1961 0.104 0.056 0.004 h0l: l=2n
9 glide plane n(b) 1961 0.100 0.056 0.004 h0l: h+l=2n
10 glide plane a(c) 1074 0.960 0.058 *** 0.991 hk0: h=2n
11 glide plane b(c) 1074 0.080 0.058 0.003 hk0: k=2n
12 glide plane n(c) 1074 0.072 0.050 0.002 hk0: h+k=2n
<!--SUMMARY_END-->
Possible spacegroups:
--------------------
Indistinguishable space groups are grouped together on successive lines
'Reindex' is the operator to convert from the input hklin frame to the standard spacegroup frame.
'SysAbsProb' is an estimate of the probability of the space group based on
the observed systematic absences.
'Conditions' are the reflection conditions (absences)
'TotProb' is a total probability estimate (unnormalised) including the probability
of the crystal being centrosymmetric from the <|E^2-1|> statistic.
Chiral space groups are marked '*' and centrosymmetric ones 'O'
Spacegroup TotProb SysAbsProb Reindex Conditions
<P n a a> ( 56) O 0.823 0.911 h00: h=2n, 0k0: k=2n, 00l: l=2n, 0kl: k+l=2n, h0l: h=2n, hk0: h=2n (zones 1,2,3,6,7,10)
---------------------------------------------------------------
Selecting space group P n a a as there is a single space group with the highest score
The spacegroup that was used for CORRECT does not matter. The next step then is to generate a HKLF 4 file, using XDSCONV:
SPACE_GROUP_NUMBER= 56 UNIT_CELL_CONSTANTS= 14.433 28.704 8.488 90.000 90.000 90.000 INPUT_FILE=XDS_ASCII.HKL OUTPUT_FILE=56.hkl SHELX
Please note that the file 56.ins has to be set up manually in this case (just take the 56.ins from above, and adjust the symops and cell parameters). The numbers after "FIND" and "PLOP" should probably be adjusted in proportion to the expected number of atoms in the asymmetric unit.
Solve the structure with SHELXD[edit | edit source]
Just run "shelxd 56". You may interrupt it with Ctrl-C once it has found a good solution, as suggested by
Try 11:20 Peaks 99 92 87 87 87 83 77 73 71 70 68 68 64 64 64 63 62 62 61 60 R = 0.294, Min.fun. = 0.747, <cos> = 0.491, Ra = 0.235 Try 11, CC All/Weak 59.81 / 46.01, best 59.81 / 46.01, best final CC 0.00 Peaklist optimization cycle 1 CC = 77.51 % BG = 0.322 for 22 atoms Peaks: 99 90 87 85 82 77 75 74 66 64 64 64 63 63 62 57 39 39 36 36 33 31 Fragments: 17 5 Peaklist optimization cycle 2 CC = 88.80 % BG = 0.225 for 25 atoms Peaks: 99 95 89 88 87 84 82 79 78 78 77 76 75 75 74 73 73 71 71 69 67 65 40 Fragments: 25 Peaklist optimization cycle 3 CC = 88.85 % BG = 0.223 for 25 atoms Peaks: 99 96 89 87 86 86 82 79 79 76 76 75 75 75 73 73 72 71 69 69 67 65 63 Fragments: 25
This solution obviously fulfills the requirement "When the final correlation coefficient CC (after PLOP) for an atomic resolution ab initio run of SHELXD is 65% or greater, the structure is almost certainly solved." in http://shelx.uni-ac.gwdg.de/SHELX/shelxdec/shelx-de.pdf .
The resulting 56.res is:
REM TRY 23 FINAL CC 88.85 TIME 3 SECS REM Fragments: 25 REM TITL 56 in Pccn CELL 0.71073 14.4330 28.7040 8.4880 90.000 90.000 90.000 ZERR 11.00 0.0029 0.0057 0.0017 0.000 0.000 0.000 LATT 1 SYMM 0.5-X, 0.5-Y, Z SYMM -X, 0.5+Y, 0.5-Z SYMM 0.5+X, -Y, 0.5-Z SFAC C UNIT 220 C001 1 0.45835 0.41566 0.09083 11.00000 0.1 99.00 C002 1 0.36894 0.55007 -0.58932 11.00000 0.1 95.84 C003 1 0.52129 0.72099 -0.95623 11.00000 0.1 89.35 C004 1 0.67521 0.30725 0.04587 11.00000 0.1 87.55 C005 1 0.40328 0.54911 -0.45947 11.00000 0.1 85.96 ... C021 1 0.60567 0.70055 -0.97749 11.00000 0.1 66.94 C022 1 0.49503 0.62079 -0.48787 11.00000 0.1 64.91 C023 1 0.60066 0.62034 -0.48599 11.00000 0.1 63.62 C024 1 0.63251 0.26331 0.06189 11.00000 0.1 63.01 C025 1 0.47217 0.73227 -1.09548 11.00000 0.1 61.79 HKLF 4 END
hints from George Sheldrick[edit | edit source]
From a November 2011 posting: UNIT specifies the number of atoms of each type in the unit-cell. For such 'small-molecule' problems you should try to get the numbers of heavier atoms correct, if only CHNO are present any numbers will do.
For such problems I recommend setting FIND to about 70% of the number of atoms (excluding H) in the asymmetric unit.
The first PLOP number should be approximately the number of atoms (excluding H) in the asymmetric unit. The second PLOP number should be about 1.2 times this and the third about 1.4 times it (three PLOP cycles are enough). This allows the 'peaklist optimization algorithm' to throw out some of the atoms.
You will need data to 1.2A or better (1.0 is much better than 1.2!). The data should be as complete as possible.
Refine using SHELXL[edit | edit source]
Copy 56.res to 56.ins. Insert
ACTA LIST 6 L.S. 10
after the UNIT 220 instruction, and run "shelxl 56". This gives a first refined model, and its electron density map, plus the relevant statistics.
general idea of refining a structure[edit | edit source]
Starting from a rough guess of the number of atoms, we adjust the model, guided by the refinement results. This is an iterative process, in which we repeatedly edit 56.res to reflect our change of conception of the structure, replace 56.ins with it, and run SHELXL again.
assigning chemical types[edit | edit source]
Since we know that there's not only carbon atoms, but likely also N, O and H, we modify 56.ins to have
SFAC C N O H UNIT 200 100 100 40
(the actual numbers after UNIT can be taken from the .lst file of SHELXL, they don't seem to matter much.)
We tell SHELXL the chemical identity by putting a 1 for a C, a 2 for a N, a 3 for an O, and a 4 for a H - the number is just the order of the atom in the SFAC line.
The chemical identity of an atom can be found from geometric parameters, and its electron density. The electron density can be displayed e.g. in coot, by loading the 56.fcf file written by SHELXL. Geometric parameters (in particular distances) are listed in the 56.lst file. Typical bond distances of C-C, C=C, C-O, C=O, C-N and X-H are about 1.54, 1.34, 1.43, 1.24, 1.47 and 1.0 A, respectively.
As a proxy to electron density we can use the refined ADPs. Atoms initially called "C", but with very low U values after refinement, are most likely O or N atoms.
Hydrogens[edit | edit source]
For the H atoms, we just move the atoms from the bottom of the .res file to those lines where the refined atoms are, if the distances to existing (heavy) atoms are close to 1 A. For hydrogens bond to C N O (and in some cases B) we could alternatively use the HFIX instruction which sets up suitable AFIX instructions for the standard 'riding H-atom' refinement (shelXle - see below - can do this with one click). This requires lines of the form
HFIX 13 XXX
In this example; the first digit 1 means tert-CH (2 would mean methylen-CH2, 3 would mean methyl-CH3, 4 would mean aromatic CH), and the second digit 3 means the normal riding model. XXX stands for the (heavy) atom name. For docs and more examples see [3].
Finishing the structure[edit | edit source]
Finally we switch to anisotropic refinement by putting an
ANIS
line into 56.ins . More info about refinement options is in the SHELXL article!
Resolution[edit | edit source]
To quote George Sheldrick:: SHELXL "prints R values for all data and for I>2sig(I) [F>4sig(F)]. The user can of course improve these by cutting back the resolution but if he or she oversteps 0.84A he/she will be caught by the CIF police. This works like a radar trap so weak datasets are usually truncated to 0.84A whether or not there are significant data to that resolution. It is always instructive to compare the R-values for all data and I>2sig(I); if the former is substantially larger, a lot of noisy outer data have been included."
Electron density[edit | edit source]
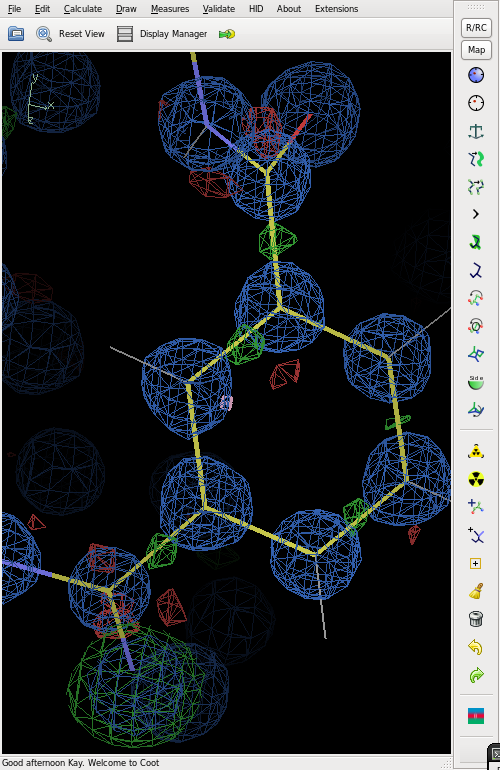
The figure shows the final electron density (blue), but with an O atom refined as N. This gives strong positive (green) difference electron density.
The difference map also shows distinct bonding electron density on most of the bonds.
A GUI for refining small molecule structures[edit | edit source]
A GUI called shelXle written by Christian Huebschle is now available for refining small molecule crystal structures with shelxl: http://ewald.ac.chemie.uni-goettingen.de/shelx