Coot: Difference between revisions
m →building loops: Fix website references |
|||
| (26 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
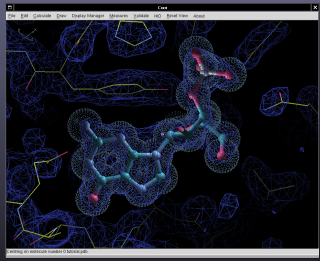
[[Image:Coot-with-ATP-vector.png| | [[Image:Coot-with-ATP-vector.png|25%|thumb|right]] | ||
Coot is a graphics program for building, refining and analysing macromolecular models obtained with crystallographic procedures. | Coot is a graphics program for building, refining and analysing macromolecular models obtained with crystallographic procedures. | ||
There is a [http://www2.mrc-lmb.cam.ac.uk/ | There is a [http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ homepage] with extensive [http://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot/web/docs/ documentation]. The program may there be [http://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot/binaries/release downloaded] for Linux and Windows. There's also Paul Emsley's [https://pemsley.github.io/coot/ coot blog]. The license of Coot is GNU GPL. | ||
=Installing Coot= | =Installing Coot= | ||
| Line 12: | Line 12: | ||
==Installing Coot on Windows== | ==Installing Coot on Windows== | ||
Please refer to | Please refer to [http://bernhardcl.github.io/coot/ Bernhard Lohkamp's WinCoot download] page. Or check manually on | ||
[https://github.com/bernhardcl/coot/releases GitHub]. | |||
==Installing Coot on Linux== | ==Installing Coot on Linux== | ||
| Line 19: | Line 20: | ||
=== Installation from a distributed binary tarball package === | === Installation from a distributed binary tarball package === | ||
This is the recommended way for those who do not want to delve into the mysteries of compiling and linking a great but complex piece of software. Read the | This is the recommended way for those who do not want to delve into the mysteries of compiling and linking a great but complex piece of software. Read the [https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ Coot documentation] to find out about the details. | ||
In short, just go to | In short, just go to https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/binaries/release/ and pick a suitable binary, e.g. | ||
coot-0. | coot-0.9.4.1-binary-Linux-x86_64-scientific-linux-7.6-python-gtk2.tar.gz for a Red Hat Enterprise Linux 7 (or CentOS-7) or newer system. | ||
Then un-tar it under /usr/local/src (or in your $HOME), and establish a symlink (ln -s) between /usr/local/bin/coot and the bin/coot of the freshly unpacked distribution. | Then un-tar it under /usr/local/src (or in your $HOME), and establish a symlink (ln -s) between /usr/local/bin/coot and the bin/coot of the freshly unpacked distribution. | ||
| Line 78: | Line 79: | ||
=== Installation from source code via autobuild scripts === | === Installation from source code via autobuild scripts === | ||
Go to https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/web/build-install-coot-from-scratch.html | |||
=Running Coot= | =Running Coot= | ||
| Line 108: | Line 87: | ||
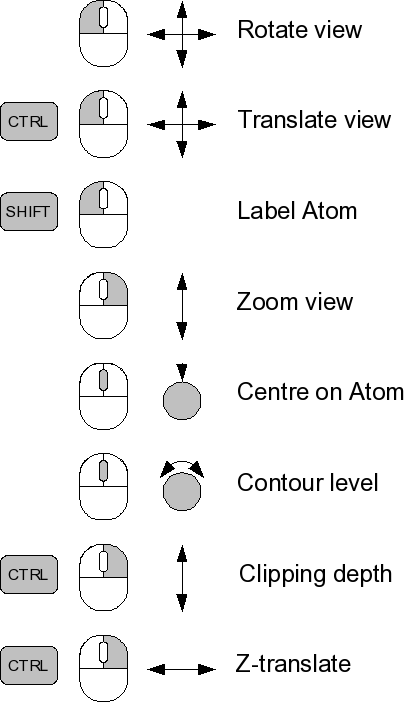
===Controls=== | ===Controls=== | ||
[[Image:Coot-controls-small.png | [[Image:Coot-controls-small.png]] | ||
| Line 126: | Line 105: | ||
[[Image:a_zalman_zm_m220w__2d_35_pic.jpg|150px|thumb|right|3d lcd]] | [[Image:a_zalman_zm_m220w__2d_35_pic.jpg|150px|thumb|right|3d lcd]] | ||
Control stereo separation / depth in coot 0.9: the transformation between the eyes is no longer a rotation and is now a shear. Which means that now we don't get part of the map showing up in the left eye but not the right (or vice versa). | |||
This is documented in Section 3.4.1, e.g. | |||
set_hardware_stereo_angle_factor(0.5) # for the Python fans | |||
Hardware stereo failure: try the line | |||
(set-display-lists-for-maps 0) | |||
in ~/.coot for coot 0.9 - see https://www.jiscmail.ac.uk/cgi-bin/webadmin?A2=ind2002&L=COOT&P=R312 and https://www.jiscmail.ac.uk/cgi-bin/wa-jisc.exe?A2=COOT;49fa8d15.2006 | |||
==== Zalman Stereo ==== | ==== Zalman Stereo ==== | ||
| Line 150: | Line 140: | ||
===External Links=== | ===External Links=== | ||
====[ | ====[https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/web/docs/coot.html On-line User Manual]==== | ||
====[ | ====[https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ Coot's home page]==== | ||
====[http://www.mail-archive.com/coot@jiscmail.ac.uk/ Current mailing list archives]==== | ====[http://www.mail-archive.com/coot@jiscmail.ac.uk/ Current mailing list archives]==== | ||
==Scheme Scripts== | ==Scheme Scripts== | ||
| Line 330: | Line 316: | ||
[[ensemble_plugin.py]] | [[ensemble_plugin.py]] | ||
===Example 6: some basic scripting=== | |||
Q: I want to prepare scripts to run coot functions like water picking or rotamer fitting in command line. Therefore I am looking for some examples of simple coot scripts, e.g. load ref.pdb, load ref.mtz, fft create map. | |||
Answer: | |||
<pre> | |||
imol = read_pdb("6vw1.pdb") | |||
imol_map = make_and_draw_map("6vw1/6vw1_refine.mtz", "2FOFCWT", "PH2FOFCWT", "", 0, 0) | |||
find_waters(imol_map, imol, 0, 1.4, 1) | |||
</pre> | |||
==Python to Scheme and return== | ==Python to Scheme and return== | ||
| Line 379: | Line 375: | ||
run_scheme_command("(scheme-command arg1 arg2 ...)") [from python] | run_scheme_command("(scheme-command arg1 arg2 ...)") [from python] | ||
=Assorted questions and answers (from the mailinglist)= | =Assorted questions and answers (from the mailinglist)= | ||
| Line 415: | Line 390: | ||
Q: Is it possible to deactivate the nomenclature errors check? Sometimes this check is not very useful and it becomes rather annoying when one has several molecules loaded only wants to look at the structures... | Q: Is it possible to deactivate the nomenclature errors check? Sometimes this check is not very useful and it becomes rather annoying when one has several molecules loaded only wants to look at the structures... | ||
A: Add to your ~/.coot | A: The [https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/web/docs/coot.html#set_002dnomenclature_002derrors_002don_002dread Coot manual] should help: Add to your startup script ~/.coot : | ||
(set-nomenclature-errors-on-read "ignore") | (set-nomenclature-errors-on-read "ignore") | ||
In case you are using the python file ~/.coot.py , use | |||
set_nomenclature_errors_on_read("ignore") | |||
On Windows, this line should go into X:\YourWinCootDirectory\.coot-preferences\coot.py . | |||
Alternatively put ".coot.py" in X:\YourWinCootDirectory. | |||
==NCS edits== | ==NCS edits== | ||
| Line 684: | Line 663: | ||
</pre> | </pre> | ||
Untried: if you have Phenix installed: it comes with phenix.probe and phenix.reduce - you could insert the paths to these binaries into the above definitions. | Untried: if you have Phenix installed: it comes with phenix.probe and phenix.reduce - you could insert the paths to these binaries into the above definitions. | ||
== some symmetry mates not shown == | |||
Q: This structure has been solved and refined using phenix in the hexagonal setting of space group R 3. There is one copy per asymmetric unit in R 3. As you can see from the attached image, coot is rendering some but not all of the symmetry mates. | |||
A: Turn up the radius a bit and use (set-symmetry-shift-search-size 3) . I would have thought that 2 is big enough, but maybe not in this case. | |||
== specify Coot download directory, like specifying backup dir by COOT_BACKUP_DIR? == | |||
here is my working startup.py (of course replace src folder with your preferred location): | |||
import os | |||
try: | |||
os.symlink('/ccp4/xray/tmp/', 'coot-download') | |||
except: | |||
pass | |||
(Eugene Osipov on 22/04/2020) | |||
== how to enable the interactive dots during RS refinement and the Ramachandran polyeders? == | |||
(asked by Clemens Grimm on 08/10/2020 ) | |||
A: | |||
set_show_intermediate_atoms_rota_markup(1) | |||
set_show_intermediate_atoms_rama_markup(1) | |||
set_do_coot_probe_dots_during_refine(1) | |||
=See also= | |||
[https://www.youtube.com/watch?v=Xhonm4K1y0c This video] is one of what will be a series of videos showcasing Coot 1 - and this one starts at the beginning. | |||
Latest revision as of 10:07, 11 January 2023

Coot is a graphics program for building, refining and analysing macromolecular models obtained with crystallographic procedures.
There is a homepage with extensive documentation. The program may there be downloaded for Linux and Windows. There's also Paul Emsley's coot blog. The license of Coot is GNU GPL.
Installing Coot
Installing Coot on OS X
OS X install packages for nightly builds that work on 10.8.X and 10.9.X are available here: Coot OS X package installers
Please refer to the Installing Coot on OS X page
Installing Coot on Windows
Please refer to Bernhard Lohkamp's WinCoot download page. Or check manually on GitHub.
Installing Coot on Linux
Installing coot on linux is rather more straightforward than on OS X, because most linux systems are based on gnome and/or kde, and tend to have many of the required components already installed. Most of the other dependencies are also readily available.
Installation from a distributed binary tarball package
This is the recommended way for those who do not want to delve into the mysteries of compiling and linking a great but complex piece of software. Read the Coot documentation to find out about the details.
In short, just go to https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/binaries/release/ and pick a suitable binary, e.g. coot-0.9.4.1-binary-Linux-x86_64-scientific-linux-7.6-python-gtk2.tar.gz for a Red Hat Enterprise Linux 7 (or CentOS-7) or newer system.
Then un-tar it under /usr/local/src (or in your $HOME), and establish a symlink (ln -s) between /usr/local/bin/coot and the bin/coot of the freshly unpacked distribution.
If you then run coot, and the loader complains that a certain library is missing, just ask
yum whatprovides <thatlibrary>
and install the library, again using yum (assuming yum is available in your distribution, otherwise use apt or whatever is there for this purpose).
Example: installing a 64bit nightly CentOS5 binary build on 64bit SL6.1
First of all, SL (Scientific Linux) is a derivative of RHEL, as is CentOS. So all three OSs behave exactly the same. The binaries with "x86_64" binaries are for 64bit systems; the "i386" binaries are for 32bit systems. Since my notebook is 64bits ("uname -a" reports "x86_64" more than once), I download coot-0.7-pre-1-revision-3999-binary-Linux-x86_64-centos-5-python-gtk2.tar.gz. As root, I "cd /usr/local/src" and un-tar. Next, have to find out which libraries are missing. This can be achieved by (note the use of LD_LIBRARY_PATH in the second command - do not permanently modify LD_LIBRARY_PATH !):
[root@localhost]# cd coot-Linux-x86_64-centos-5-gtk2-python [root@localhost]# LD_LIBRARY_PATH=lib ldd bin/coot-real | grep found libssl.so.6 => not found libcrypto.so.6 => not found libssl.so.6 => not found libcrypto.so.6 => not found
So only two libraries are missing! Either they can be installed using yum, or they are already available, but have a higher version.
- First possibility: find out about installable RPM packages (preferred way):
[root@localhost src]# yum provides libssl.so.6 libcrypto.so.6
Loaded plugins: refresh-packagekit
openssl098e-0.9.8e-17.el6.i686 : A compatibility version of a general
: cryptography and TLS library
Repo : sl
Matched from:
Other : libssl.so.6
... (the package is repeated, and libcrypto.so.6 is also mentioned)
- Now don't just install the openssl098e-0.9.8e-17.el6.i686 and its dependencies - it is a 32bit library (the name ends with ".i686")! Installing it does not solve the problem - we need a 64bit library. Unfortunately "yum provides" does not tell us about the 64bit library (is that a yum bug?). By specifying just the package name (openssl098e.x86_64 would also work, and would avoid any 32bit package)
yum install openssl098e
- we install both libssl.so.6 and libcrypto.so.6 in their 64bit versions - done!
- Second possibility: find out if the system already has a higher version of the two libraries:
[root@localhost locate libssl.so /usr/lib64/.libssl.so.1.0.0.hmac /usr/lib64/.libssl.so.10.hmac /usr/lib64/libssl.so /usr/lib64/libssl.so.1.0.0 /usr/lib64/libssl.so.10
- So the answer is: there is /usr/lib64/libssl.so which is at version 10, which is compatible with the version we need (6). For libcrypto.so the same is true. So just
cd coot-Linux-x86_64-centos-5-gtk2-python/lib/ ln -s /usr/lib64/libssl.so libssl.so.6 ln -s /usr/lib64/libcrypto.so libcrypto.so.6
- The way these symlinks are made they would even work if RHEL upgrades libssl or libcrypto to higher versions. Works for me.
Final step (this does not need to be repeated for a new coot version): create /usr/local/bin/coot with
#!/bin/csh -f setenv LANG C exec /usr/local/src/coot-Linux-x86_64-centos-5-gtk2-python/bin/coot $*
and make it executable with
chmod a+x /usr/local/bin/coot
Installation from source code via autobuild scripts
Go to https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/web/build-install-coot-from-scratch.html
Running Coot
General Topics
Controls
Stereographic Display
Coot has several options for stereographic display, ranging from cross-eyed and wall-eyed split-screen stereo, to hardware-stereo modes that work with CRT systems and most recently the new Zalman 3-D LCD monitor.
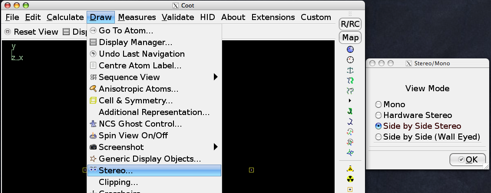
Side-by-Side
Either cross-eyed or wall-eyed split-screen stereo mode can be invoked using the "Stereo" menu item under "Draw", as is shown in the image below:
Hardware Stereo
Similarly, hardware stereo can be invoked (assuming you have the CRT, correct graphics card, emitter, etc) using the same menu item, by selecting "Hardware Stereo".

Control stereo separation / depth in coot 0.9: the transformation between the eyes is no longer a rotation and is now a shear. Which means that now we don't get part of the map showing up in the left eye but not the right (or vice versa).
This is documented in Section 3.4.1, e.g.
set_hardware_stereo_angle_factor(0.5) # for the Python fans
Hardware stereo failure: try the line
(set-display-lists-for-maps 0)
in ~/.coot for coot 0.9 - see https://www.jiscmail.ac.uk/cgi-bin/webadmin?A2=ind2002&L=COOT&P=R312 and https://www.jiscmail.ac.uk/cgi-bin/wa-jisc.exe?A2=COOT;49fa8d15.2006
Zalman Stereo
The first viable LCD monitor for stereographics display is made by Zalman and costs about $300: Zalman ZM-M220W
The attributes for this monitor have been tested and described rather extensively by Warren DeLano on the PyMOL site. Please read it for important details and suggested purchasing sources.
The coot zalman page describes specifically how to get this to work with coot on Mac OS X, but the instructions should be generalizable to linux and Windoze.
Note that the stereo effect is very sensitive to the vertical position of your eyes relative to the screen: if you don't see stereo, try tilting the screen.
Stereo: left/right (and front/back) interchanged?
Establish an additional toolbutton "swap stereo":
Main Toolbar -> right mouse click-> Manage buttons-> select Swap Stereo
Or for the script minded:
switch_stereo_sides()
This will toggle the stereo images left and right.
External Links
On-line User Manual
Coot's home page
Current mailing list archives
Scheme Scripts
Coot can be scripted in scheme (guile) or python - support for each is more or less equal these days.
Several examples of coot extensions to the language can be seen by examining the 0-coot-state.scm file that coot leaves behind when it finishes.
COOPS
Coops generates a coot script from the output of molprobity, specifically probe, reduce, cluster and clashlist.
For an explanation of the principals underlying reduce and clashlist see the Dots Page. Get Molprobity software here.
Use Coot version 0.1 or higher.
Invoke like this (from the directory in which you run coot):
$ coops myfile.pdb
The use Calculate->Scripting to read in and run coops.scm
Get COOPS here.
Example Scheme Script 1: Move to Molecule Centres
This example can be found in the coot scheme sources (the function name is molecule-centres-gui and is in the xxx/share/coot/scheme/coot-gui.scm file). It is a simple function that creates a button box - a button for each coordinates molecule in Coot. It is annotated. Reproduced as coot-scheme1.
Example Scheme Script 2: Demo a Few of Coot's Features
Reading in a pdb file, an MTZ file and manipulating the model
This is a composite script and demonstrate reading pdb file, an MTZ file, translations, zoom, spin zooms, contour level changing, map masking, real space refinement, water addition and loop fitting.
The data files used in the example can be obtained here and here. Put them in the directory where you start coot. Save the script to your disk, then use Calculate -> Run Script... to activate it.
Example Scheme Script 3: Read CNS data
This CNS data reading script is a Cootenization of the CN2COOT script written by Joel Bard (it is based on his csh script) and can be used to compare and contrast scheme programming and shell script programming (the coot version is longer to some extent because it does extra error checking).
As well as doing the conversion the resulting mtz/maps are loaded into Coot.
It is part of Coot as of version 0.1.2.
Example Scheme Script 4: Load the Latest Data and PDB files Automatically
To load the most recent files, do this:
coot --script latest-files.scm
which enables the scripting function: (load-latest-files)
For extra gui goodness (you will need 0.1.2):
coot --script latest-files.scm --script extensions.scm
Example Scheme Script 5: Saving a Partial model
Here we create a small function to save part of a molecule and add a gui interface, it can be used in the usual way (i.e. with --script on the command line, Calculate->Run Script... or add the script to your ~/.coot file.
save-partial.scm
Example Scheme Script 6: Creating an interface for the Powermate Dial
The Powermate dial can be used with coot. One could just assign the rotations to +/-y keys and be done with it, but this script gives you a way of having positive and negative rotations in all three cartesian directions. The F1 key is mapped to positive rotation, the F2 key to negative rotation, and the F3 key permits you to toggle through x, y, and z, on successive key presses. I then map F1 and F2 into the ordinary rotations on the powermate (using send key equivalents) and then I map F3 into the single click on the dial, making it easy to toggle through x, y and z. The press-and-rotate options remain available; I map these into scroll up and down, and put them on the slowest response setting, which makes contouring density easier to control than it is from my mouse scroll wheel.
powermate-coot.scm
Example Scheme Script 7: Applying arbitrary value to "B" factor column
Imagine you have a file of some property (Chemical Shifts, for example) of a residue that you wish to apply to the atoms of a particular model from a pdb file as pseudo B factors. Here's how to do that in Coot:
We have a file "cs.tab" like this, the residue number then the chemical shift value (one for each residue in a particular chain):
1 1.53159
2 4.35884
3 4.07123
4 4.16932
5 6.69103
6 7.12071
7 10.7419
8 9.57176
Use apply-cs.scm to apply these values as pseudo temperature factors. Typical usage, where "A" is the chain id, and cs.tab the file of values per residue.
(apply-cs (read-pdb "test.pdb") "A" "cs.tab")
--script apply-cs.scm
Example Script 8: Partial Occupancy Dialog
Imagine that you have a structure that has residues with partial occupancy. After refinement, it would be convenient to quickly navigate to all such residues. How can that be done?
Start coot with command line arguments:
--script partial-occupancy-navigation.scm
This will provide an extra menu item called "Extras", clicking on "Residues with low occupancy..." therein will lead you through the process. Note that this will often work with SHELXL molecules, because they have atoms with negative (e.g -31) occupancies.
Note also that you will need a recent version of Coot to use this, as it stands. This will not work on stock Coot version 0.4.x. You can enable this for use with 0.4.x if you update/replace your xxx/share/coot/scheme/coot-gui.scm file from here.
Example Script 9: A GUI for Chopping Back Sidechains from a Residue Range
This is a simple interface to the delete-sidechain-range function, it illustrates how arguments can be transfered from the GUI to the scripting function. It was written in response to a question from Byron DeLaBarre.
Unfortunately (prior to 0.5) there was an error in the standard delete-sidechain-range function, which is why we over-ride it.
--script chop-side-chains-gui.scm
Example 10: How do I bind a key to Toggle the display of NCS ghosts?
With this script: toggle-ncs-ghosts-script
Example 11: Paul Emsley's Key Bindings
Just so you get an idea of the customization by key bindings here are what Paul uses currently (add to your .coot file).
Optional Wrappers and (External) Shell Script Enhancements
I (wgscott) wrote a coot wrapper shell script that lets you convert xplor/cns maps on the fly (you need to install mapman first) and has a few other enhancements.
I also made a Coot OS X applet that allows you to drag and drop a cns/xplor or ccp4 mapfile or any other coot-compatable file (mtz or pdb file, for example). Using the File > Get Info dialog, you can program this applet to open all .map and all .mtz files, if you want to, making these files double-clickable.
Download the Applet (requires a separate working coot installation)
Python Scripts
Example 1: Bernhard Lohkamp's Key Bindings
Just so you get an idea of the customization by key bindings here are what Bernhard/Paul uses currently (add to your .coot file or put the file in .coot-preferences directory).
bernhards_key_bindings_for_coot.py
Example 2: More key bindings (inspired by the Coot BB)
For (re-)colouring maps blue:
To (re-)colour coordinate molecules yellow:
Example 3: NCS Rotamer differences
To show NCS where NCS-related side-chains have different rotamers:
Example 4: Morphing GUI
GUI to easily access jiggle fit and morphing (currently pre-release Coot required, may be moved into trunk):
Example 5: Ensemble GUI
GUI to allow navigation through structural ensembles as obtained e.g. from ensemble refinement:
Example 6: some basic scripting
Q: I want to prepare scripts to run coot functions like water picking or rotamer fitting in command line. Therefore I am looking for some examples of simple coot scripts, e.g. load ref.pdb, load ref.mtz, fft create map.
Answer:
imol = read_pdb("6vw1.pdb")
imol_map = make_and_draw_map("6vw1/6vw1_refine.mtz", "2FOFCWT", "PH2FOFCWT", "", 0, 0)
find_waters(imol_map, imol, 0, 1.4, 1)
Python to Scheme and return
Translating between Python and Scheme
Python scripting is different to (default) scheme scripting which is mainly described in Paul Emsley's documentation (although it's mentioned somewhere, fairly hidden). You have to change the commands in the following way:
GUILE scripting: (guile-command argument1 argument2 ...)
PYTHON scripting: python_command(argument1, argument2, ...)
Simple rules for Scheme to Python translations
Here some simple rules how to translate from Scheme to Python. To translate the other way around, i.e. Python to Scheme, just turn the rules around:
- Replace all '-' with '_' (except in equation when you need arithmetic '-' minus signs)
- Move the brackets around the argument(s)
- Separate multiple arguments by commas rather than spaces
- Replace 'define' with 'def' for functions and with '=' for assignments
- Make sure to use indentation for the function content [Python is indentation sensitive] and a ':' after the function definition.
Some additional/advanced(?) rules:
- #f -> False
- #t -> True
- (set! variable value) -> variable=value
A simple example
In Scheme we may have the following script:
(define mol2-pdbFile "somePDBfile.pdb" )
(define mol2-model (read-pdb mol2-pdbFile))
(define (read-mol-again)
(clear-and-update-model-molecule-from-file mol2-model mol2-pdbFile))
(read-mol-again)
Which will translate into Python:
mol2_pdbFile = "somePDBfile.pdb"
mol2_model = read_pdb(mol2_pdbFile)
def read_mol_again():
clear_and_update_model_molecule_from_file(mol2_model, mol2_pdbFile)
read_mol_again()
Running a Scheme/Python command from Python/Scheme
As of Coot 0.5 (and if you have both scripting languages available) you an use the following commands to run a script or command in the other language:
(run-python-command "python_command(arg1, arg2, ...)") [from guile/scheme]
run_scheme_command("(scheme-command arg1 arg2 ...)") [from python]
Assorted questions and answers (from the mailinglist)
It should be noted that the answers ("A") are from Paul Emsley himself (and were maybe slightly edited).
Coot development
Q: How can I get involved with Coot development?
A: Join the Coot Janitors project. This is a project to get new people involved in improving Coot, by acting as a clearing house for simple tasks which need doing, and providing documentation for doing them.
Get rid of the "fix nomenclature" check
Q: Is it possible to deactivate the nomenclature errors check? Sometimes this check is not very useful and it becomes rather annoying when one has several molecules loaded only wants to look at the structures...
A: The Coot manual should help: Add to your startup script ~/.coot :
(set-nomenclature-errors-on-read "ignore")
In case you are using the python file ~/.coot.py , use
set_nomenclature_errors_on_read("ignore")
On Windows, this line should go into X:\YourWinCootDirectory\.coot-preferences\coot.py . Alternatively put ".coot.py" in X:\YourWinCootDirectory.
NCS edits
Q: I am sure this exists somewhere through scripting in COOT, but can I apply NCS edits to only a subset of NCS copies? In other words, can I tell coot which are NCS related chains, and which aren't. I am working on this nightmarish case of asymmetrical homodimers, where the sequences are very similar, but the structures are not, so I need to tell coot which chains are actually related to each other.
A: Nightmare. If you have a recent [1632 or later for the scheme version, 1646 for the python version] Coot, you can do this:
(manual-ncs-ghosts imol resno-start resno-end chain-id-list)
where
- imol is 0 (say)
- resno-start and resno-end is the residue range for the LSQ fitting to return the NCS matrix,
- chain-id-list is the list of chain-ids, starting with the master/reference chain-id and followed by the peer chain-ids that are NCS related, e.g. (list "A" "B" "D")
The python interface is similar.
There is also a GUI to activate this feature under Extensions -> NCS.
SHELXL
Description of problematic situation: I am using SHELXL to refine my 1.2 Å data and I am refining the hydrogen atoms. Subsequent rebuilding in coot is difficult though since hydrogens often does not "follow" when you do side chain rebuilding. For the moment I have quit transfering hydrogens to coot and add the hydrogens every refinement cycle, though it would be good I think if I could see them in coot without bothering about wrong positions. So these are my specific questions:
Specific Q1: Using "edit chi angles" does not work properly.
A: This fails because for chi angles Coot uses the Refmac dictionary to know what is connected to what (if it can). The work-around is to rename the refmac dictionary so Coot can't find it - which will force Coot to find bond by distance criteria.
Specific Q2: Using "real space refine" does not work properly.
A: Yes this fails. Hydrogens are named differently to SHELX hydrogens. In principal this could be made to work if the dictionary was reworked to use SHELX hydrogen names. This would also fix the chi angles problem too of course.
Specific Q3: I am unable to open the output pdb file from ShelXL in Coot.
A: Well, it's hard to know what's the problem without details - the console should say something. But when handling the output of shelxl, I suggest you read the .res file rather than the pdb, then the subsequent .ins file contains lots of "header" information.
Another answer to questions 1+2 is to rename the hydrogen atoms in the shelxl res-file to match the mmCIF dictionaries used by Coot. This only needs to be done once as shelxl does not modify these names. Except for a few manual editions, the renaming can be done semi-automatically using regular expressions (replacing A->1, B->2, etc).
Concerning question 3, the Coot -> Extensions -> Module -> SHELXL menu entry works really well now. It reads in all relevant shelxl files and provides a menu highlighting the problematic areas in the model.
Image quality on NVidia cards
Q: improvement of image quality on machines with NVidia cards?
A: After talking about antialiasing with Stuart McNicholas, I discovered a program called nvidia-settings. I found that if I do:
Antialiasing Settings -> Override Application Setting, and set the slider to 16x
then I start Coot, I see that it starts up in nice antialiasing mode (which is a lot better that the poor mode that Coot has built-in). I don't know if this works on other/newer systems, but it works for me on my oldish GeForce 6600. (Of course the FPS takes a hit.)
Q: How to set the default to display symmetry related molecules?
A: Add (set-show-symmetry-master 1) to the appropriate file.
Startup files
Q: I still have a ".coot" file in my home folder for a few coot preferences that I couldn't find in the new ".coot-preferences/coot-preferences.scm". There is a warning that I should not add commands to this file. So is a "~/.coot" still the proper place to add default commands for coot?
A: Coot does not create a ~/.coot file for you, but will read it if it exists. Likewise, ~/.coot.py in which you can write python commands.~/.coot-preferences is a directory in which all .scm files and .py files are executed. coot-preferences.scm and coot_preferences.py there are generated by using the Edit -> Preferences dialog (and thus it overwrites older versions - hence the warning).
If you want to create a script that will be read by everyone then put those files into a directory defined by environment variable $COOT_SCHEME_EXTRAS_DIR (for *.scm) or $COOT_PYTHON_EXTRAS_DIR (*.py). All *.scm in $COOT_SCHEME_EXTRAS_DIR and *.py files in $COOT_PYTHON_EXTRAS_DIR will be executed at start up.
So you have a variety of places. Personally I mostly use ~/.coot.
Torsion general
Q: How do I use "torsion general"?
A: Thanks for pointing out the lack of documentation on this. I'll make a note to add some.
You need to click on the torsion-general icon, then click 4 atoms that describe the torsion - the first atom will be the base (non moving) part of the atom tree, on clicking the 4th atom a dialog will pop up with a "Reverse" button [1].
Move this dialog out of the way and then left mouse click and drag in the main window will rotate the moving/"top" part of the residue round the clicked atoms 2 and 3. When you are happy, click "Accept".
Window focus may be an issue - depending on your setting, the window manager may eat one of your clicks as you change focus between the dialog and the main graphics window (this I find annoying and there are instructions in the FAQ on how to turn that off for various systems).
[1] which may not work in 0.6-pre (grumble/sigh/sorry). If it doesn't not work, the "Reverse" button should invert the moving and "base" part of the residue.
Peak heights in maps
Q: I have some peaks in my map which take water or sodium/magnesium or chlorine atom with out giving out any positive or negative density upon further refinement. Is there any easy way of calculating the peak height / number of electrons at a given position, say a mouse click point in coot? Is there any formula to calculate the number of electrons based on sigma level and peak height, as given in difference map peaks in coot?
A: First, go to the Coot wiki and pick up the scheme key bindings.
If you want density information at a given cursor point: point at the blob, press the 'g' key (which recentres on the biggest density under the cursor).
using the Scheme scripting window:
(apply density-at-point (imol-refinement-map) (rotation-centre))
There is no user access to the peak integration code of coot as yet.
Disulfide bond across crystallographic axis
Q: I have a pair of disulfide bonds which link two monomers in separate asymmetric units. There is a single monomer in the asymmetric unit, and two monomers come together to form disulfides between Cys 26-Cys45, and Cys45-Cys26. When I real-space-refine these residues, they do not form a nice disulfide, and Coot does not seem to recognize them as a disulfide.
A: For the record, you can't refine symmetry-related disulfides in Coot (as of Nov 3, 2009).
Macros in COOT
Q: How to use macros in COOT? Do they need to be written in Python or another language that I had not heard of before? Where can I find a low level description of how to write macros with some examples (I know nothing about Python, except that it is fashionable)?
A: The other language is a form of Lisp, called Scheme. You can learn about programming python in many ways of course (not least the python tutorial, which is what I read first). The coot python extensions are described in the documentation. There is a standard trivial formatting change that has to be made to get the syntax right for python, see "Python Scripting" [[1]]. There is a growing collection of coot scripts in this Wiki article.
building loops
Q: Is there any similar function in COOT as lego_auto_mainchain command in O program?
A: there are 2 loop fitting tools in Coot
- C alpha -> Mainchain [2],[3]
- DB Loop: (No good documentation) [4] Extensions -> Modelling -> DB Loop...
LSQ superpositions
Q: Do an LSQ superposition using specified residues in multiple chains (superposing one oligomer on another).
A: Something like this then?
clear_lsq_matches() # specs for reference then moving add_lsq_match(20, 90, "A", 20, 90, "A", 1) add_lsq_match(20, 90, "B", 20, 90, "B", 1) add_lsq_match(15, 75, "D", 15, 75, "D", 1) apply_lsq_matches(1, 2)
which presumes that the reference molecule is in 1 and the moving molecule 2.
Q: How to do a LSQ superposition of a homologous structure onto my working structure using ± N residues about the current position, where N is a variable (not essential, could be fixed) and the current position is the last residue that I clicked on.
A: That is more involved - and more useful because it can be dynamic. Something like the following perhaps (in Scheme, just for amusement (not tested)). You will need to set imol-ref, perhaps by reading in the reference pdb, as demonstrated below. The function is bound to Shift-Y.
(define dynamic-lsq-range-extent 2) ;; ± 2 residues either side of centre residue
(define imol-ref (read-pdb "reference.pdb"))
;; convert between the input reference chain id and the chain id of
;; the moving molecule that corresponds to that chain
;;
(define (mov-match-chain ref-chain-id)
ref-chain-id)
(define (dynamic-lsq-match)
;; get the current residue and use that to make residue ranges for
;; an LSQ fit
;;
(using-active-atom
(clear-lsq-matches)
(add-lsq-match (- aa-res-no dynamic-lsq-range-extent)
(+ aa-res-no dynamic-lsq-range-extent)
aa-chain-id
(- aa-res-no dynamic-lsq-range-extent)
(+ aa-res-no dynamic-lsq-range-extent)
(mov-match-chain aa-chain-id)
1)
(apply-lsq-matches aa-imol imol-ref)))
(add-key-binding "Dynamic LSQ overlay" "Y" dynamic-lsq-match)
reading MTZ file with experimental PHI and FOM using --auto
Q: There is the --auto <filename> commandline option for auto-reading mtz files (mtz file has the default labels FWT, PHWT). Can this be made to work with a SHELXE .phs output file after converting with convert2mtz ? - the resulting MTZ file has labels F PHI FOM.
A: use: coot --python -c 'make_and_draw_map("sad.mtz", "F", "PHI", "FOM", "/HKL_base/HKL_base/FOM",1, 0)'
NCS Rotamer differences
Show me where NCS-related side-chains have different rotamers
(define (compare-ncs-rotamer imol chain-A chain-B)
(let ((n-residues (chain-n-residues chain-A imol))
(mismatched-rotamers '()))
(for-each
(lambda (serial-number)
(let ((res-name-A (resname-from-serial-number imol chain-A serial-number))
(res-no-A (seqnum-from-serial-number imol chain-A serial-number))
(ins-code-A (insertion-code-from-serial-number imol chain-A serial-number))
(res-name-B (resname-from-serial-number imol chain-A serial-number))
(res-no-B (seqnum-from-serial-number imol chain-A serial-number))
(ins-code-B (insertion-code-from-serial-number imol chain-A serial-number)))
(if (not (= res-no-A res-no-B))
(begin
(format #t "sequence number for ~s do not match~%" res-no-A))
(if (not (string=? res-name-A res-name-B))
(begin
(format #t "residue names for ~s do not match~%" res-no-A))
(let ((rot-name-A (get-rotamer-name imol chain-A res-no-A ins-code-A))
(rot-name-B (get-rotamer-name imol chain-B res-no-B ins-code-B)))
(if (not (string=? rot-name-A rot-name-B))
(begin
(set! mismatched-rotamers
(cons (list imol chain-A res-no-A ins-code-A
res-name-A
(if (string=? rot-name-A "") "-" rot-name-A)
(if (string=? rot-name-B "") "-" rot-name-B))
mismatched-rotamers))))
)))))
(range n-residues))
(dialog-box-of-buttons "Mismatched Rotamers"
(cons 300 300)
(map (lambda(rotamer)
(let ((label (string-append " "
(list-ref rotamer 1)
" "
(number->string (list-ref rotamer 2))
(list-ref rotamer 3)
" "
(list-ref rotamer 4) ;; res-name
": "
(list-ref rotamer 5)
" vs. "
(list-ref rotamer 6)))
(thunk (lambda ()
(set-go-to-atom-molecule imol)
(set-go-to-atom-chain-residue-atom-name
(list-ref rotamer 1)
(list-ref rotamer 2) " CA "))))
(list label thunk)))
mismatched-rotamers)
" Close ")))
And one would use this something like:
;; example usage: (let ((imol (read-pdb "test.pdb"))) (compare-ncs-rotamer imol "A" "B"))
make RSR in coot 0.8.1 behave like in earlier versions
Q: We've noticed a new behavior in real space refinement in coot 0.8.1 whereby dragged atoms are more tightly restrained to their initial positions than in earlier versions. This seems to be described in the release notes by:
o BUG-FIX: The amount that the other atoms ove with moving the picked atom has been reduced (but is configurable)
A: Add e.g. this to your ~/.coot.py file:
set_refinement_drag_elasticity(0.8)
Q: I'm wondering why this was changed. Does the optimum elasticity change with resolution, map quality, or another experimental limitation? Or does it more of a user preference?
A: Because of cis-peptides. My worry was that in the previous regime, it was too easy to introduce cis-peptides when fitting to low resolution maps. I believe the current default setting is much less likely to do that.
Q: I've tried various settings of refinement_drag_elasticity and I need to lower it to 0.5 or so before any semblance of earlier behavior appears.
A: It used to be 0.167, I think.
Molprobity not active in COOT
Q: I am using COOT 0.8.1 EL that comes with the CCP4 6.5.010 on my Mac OS X 10.10.2. I wanted to run molprobity but the Validate > Probe clashes button in my pull down menu is not active. Is this function available in this COOT version?
A: Reduce and probe are separate programs available from the Richardson’s lab at Duke http://kinemage.biochem.duke.edu/. Download and install on your box. Then coot needs to be told in some instances where it can find these executables. I have the following lines in my ~/.coot file in Linux.
;; .coot ;; This file is required. As of coot 0.8pre no other mechanism for ;; enabling probe in coot works ;; ;; This is full pathname of molprobity's probe program (define *probe-command* "/apps/xray/bin/probe") ;; This is full pathname of molprobity's reduce program (define *reduce-command* "/apps/xray/bin/reduce")
Untried: if you have Phenix installed: it comes with phenix.probe and phenix.reduce - you could insert the paths to these binaries into the above definitions.
some symmetry mates not shown
Q: This structure has been solved and refined using phenix in the hexagonal setting of space group R 3. There is one copy per asymmetric unit in R 3. As you can see from the attached image, coot is rendering some but not all of the symmetry mates.
A: Turn up the radius a bit and use (set-symmetry-shift-search-size 3) . I would have thought that 2 is big enough, but maybe not in this case.
specify Coot download directory, like specifying backup dir by COOT_BACKUP_DIR?
here is my working startup.py (of course replace src folder with your preferred location):
import os
try:
os.symlink('/ccp4/xray/tmp/', 'coot-download')
except:
pass
(Eugene Osipov on 22/04/2020)
how to enable the interactive dots during RS refinement and the Ramachandran polyeders?
(asked by Clemens Grimm on 08/10/2020 )
A:
set_show_intermediate_atoms_rota_markup(1) set_show_intermediate_atoms_rama_markup(1) set_do_coot_probe_dots_during_refine(1)
See also
This video is one of what will be a series of videos showcasing Coot 1 - and this one starts at the beginning.