Examples of electron density: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
|||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== Figures from Bernhard Rupp's book "Biomolecular Crystallography" (with permission by the author) == | == Figures from Bernhard Rupp's book "Biomolecular Crystallography" (with permission by the author) == | ||
There is more material (all figures, citations, tutorials) at the [http://www.ruppweb.org/garland/ book's overview page]. | |||
You can see the full-size, high-resolution figure by clicking on it. | You can see the full-size, high-resolution figure by clicking on it. | ||
| Line 5: | Line 7: | ||
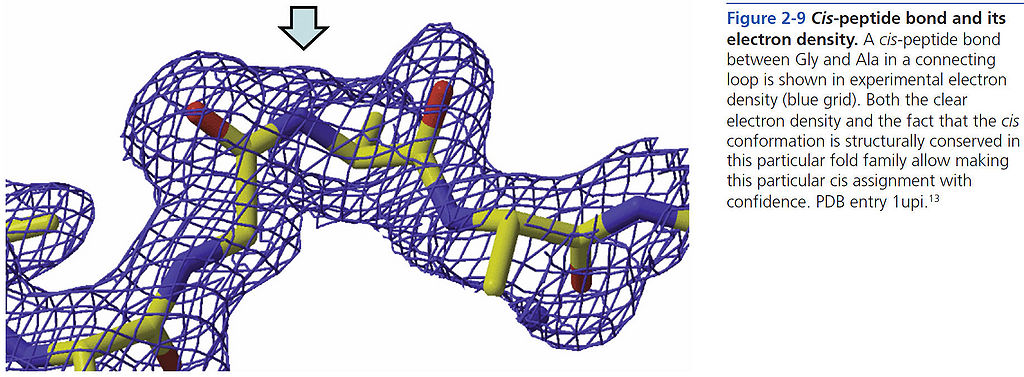
=== Cis-peptide bond and its electron density === | === Cis-peptide bond and its electron density === | ||
[[File:BMC 2-9.jpg|1024px]] | [[File:BMC 2-9.jpg|1024px]] | ||
---- | |||
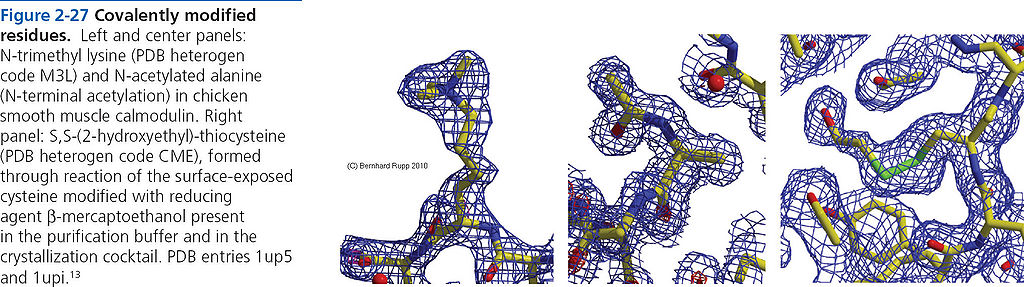
=== Covalently modified residues === | === Covalently modified residues === | ||
[[File:BMC 2-27.JPG|1024px]] | [[File:BMC 2-27.JPG|1024px]] | ||
---- | |||
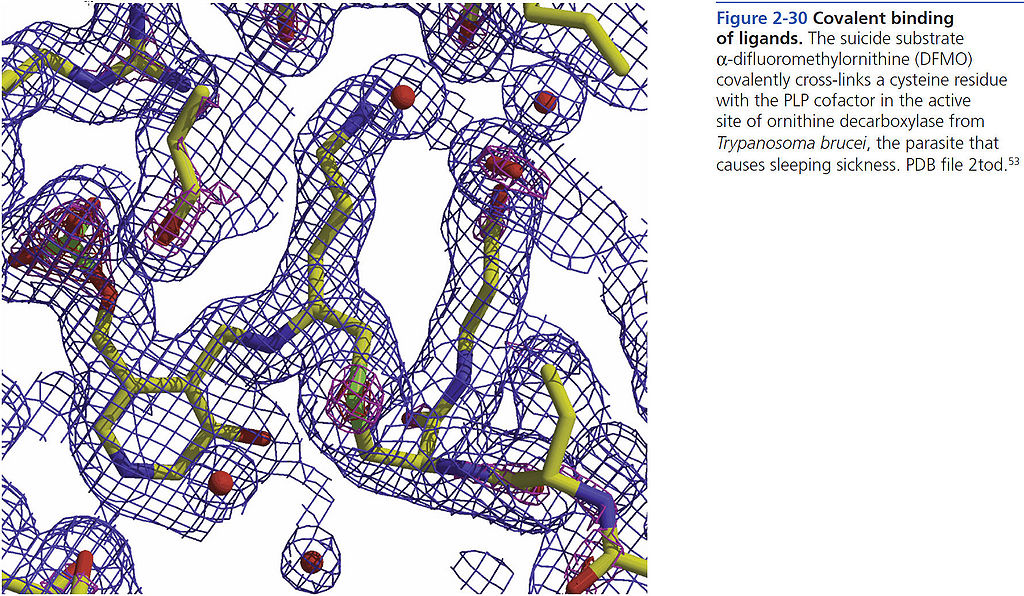
=== Covalent binding of ligands === | === Covalent binding of ligands === | ||
[[File:BMC 2-30.jpg|1024px]] | [[File:BMC 2-30.jpg|1024px]] | ||
---- | |||
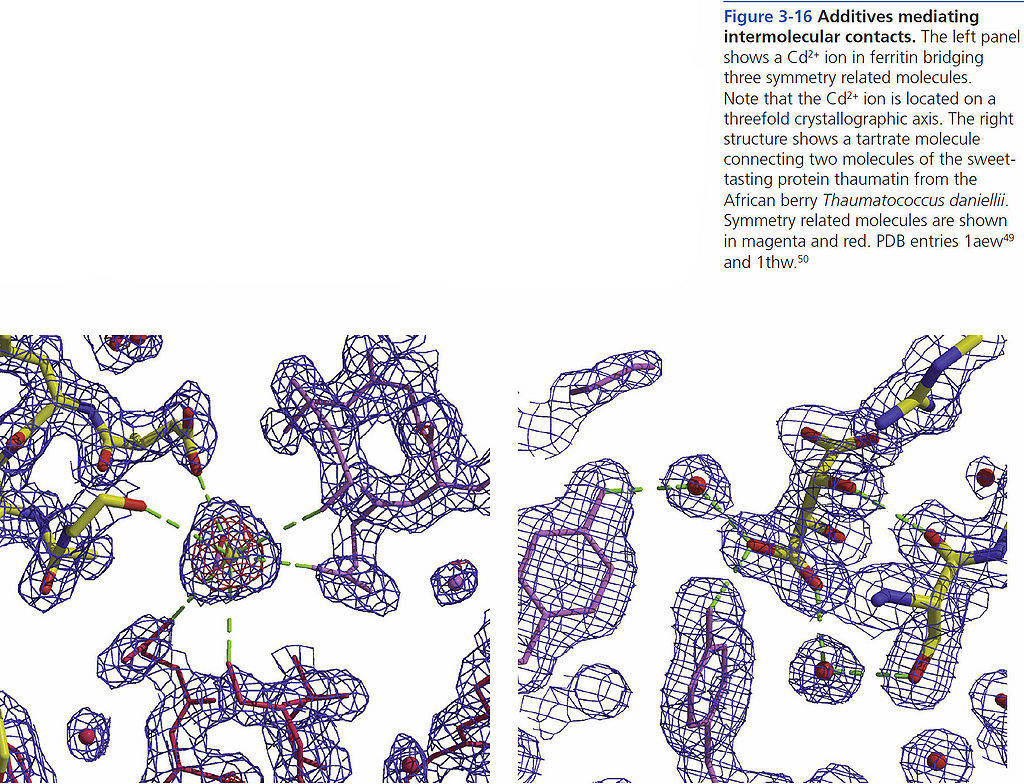
=== Additives mediating intermolecular contacts === | === Additives mediating intermolecular contacts === | ||
| Line 20: | Line 28: | ||
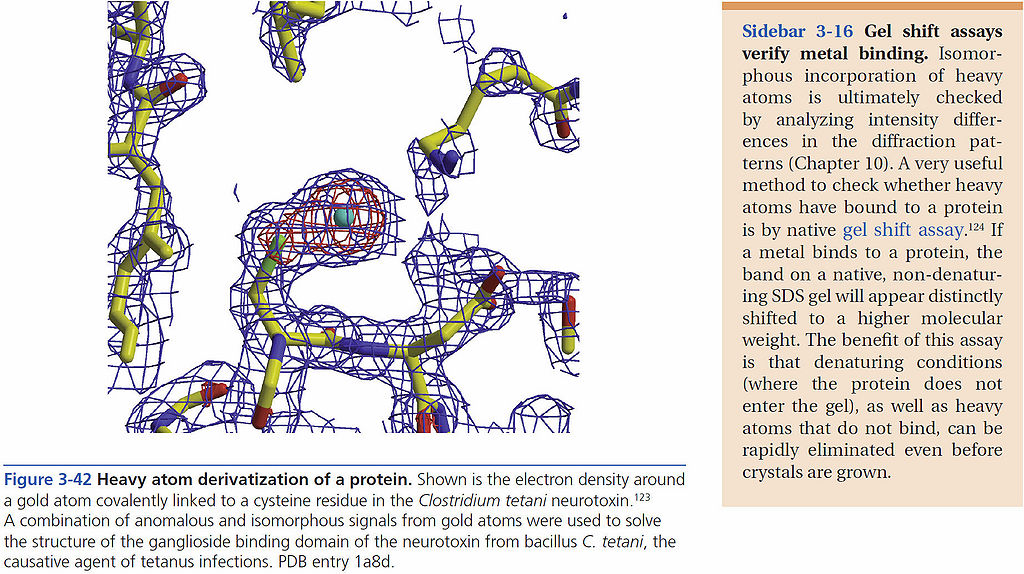
=== Heavy-atom derivatization of a protein === | === Heavy-atom derivatization of a protein === | ||
[[File:BMC 3-42.jpg|1024px]] | [[File:BMC 3-42.jpg|1024px]] | ||
---- | |||
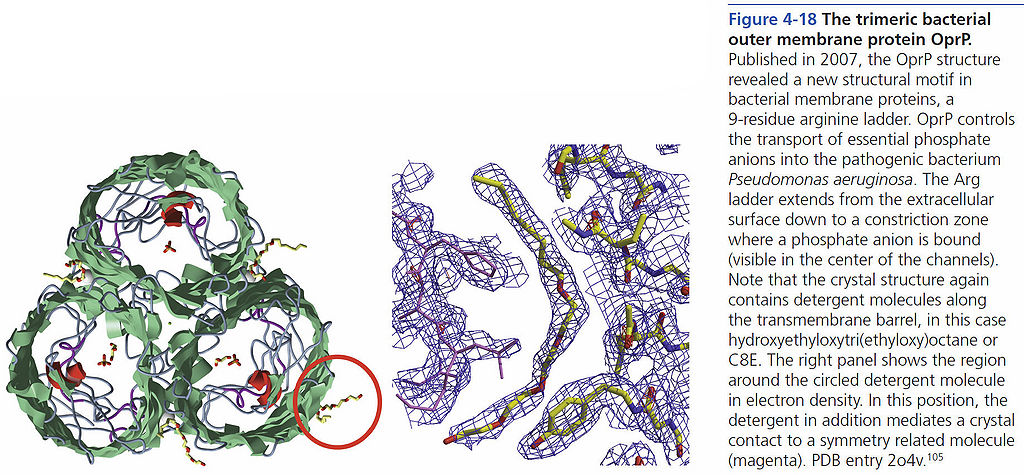
=== The trimeric bacterial outer membrane protein OprP === | === The trimeric bacterial outer membrane protein OprP === | ||
| Line 28: | Line 38: | ||
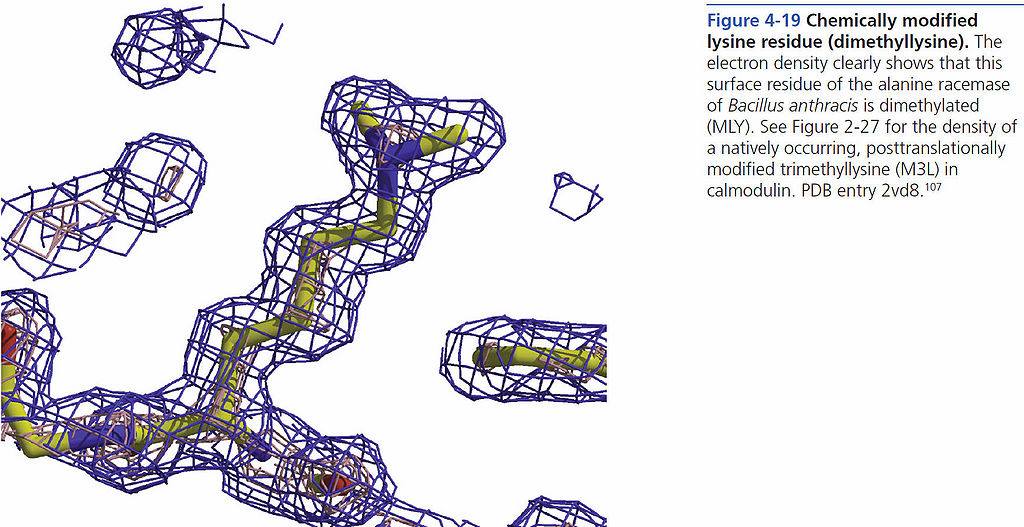
[[File:BMC 4-19.jpg|1024px]] | [[File:BMC 4-19.jpg|1024px]] | ||
---- | |||
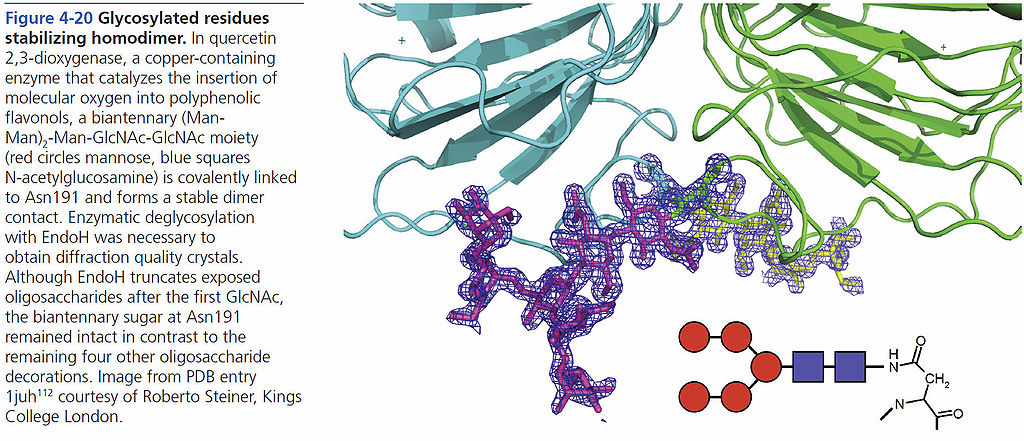
=== Glycosylated residues stabilizing homodimer === | === Glycosylated residues stabilizing homodimer === | ||
[[File:BMC 4-20.jpg|1024px]] | [[File:BMC 4-20.jpg|1024px]] | ||
---- | |||
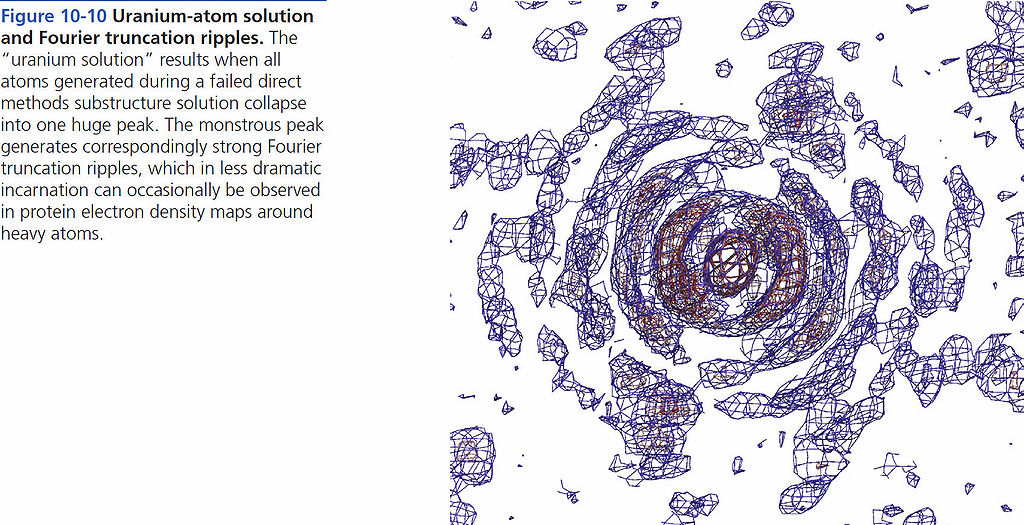
=== Uranium-atom solution and Fourier truncation ripples === | === Uranium-atom solution and Fourier truncation ripples === | ||
[[File:BMC 10-10.jpg|1024px]] | [[File:BMC 10-10.jpg|1024px]] | ||
---- | |||
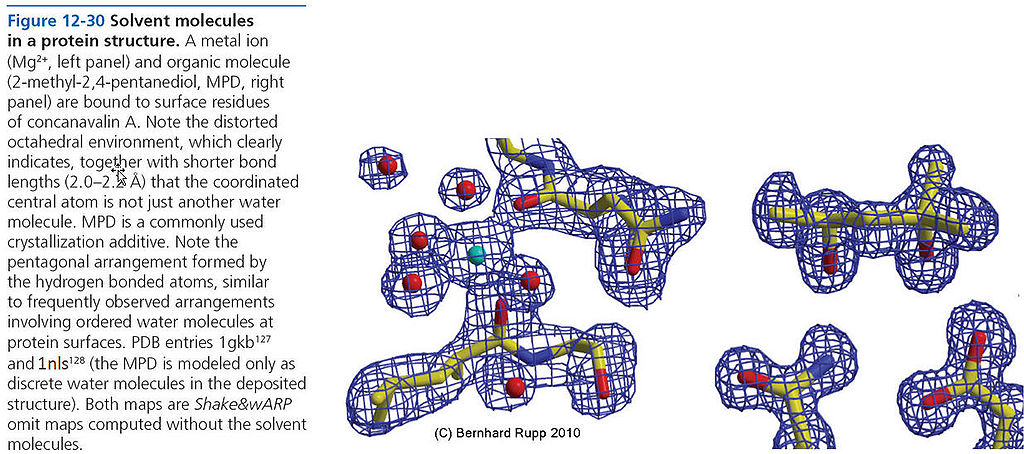
=== Solvent molecules (water, MPD, Mg2+) === | === Solvent molecules (water, MPD, Mg2+) === | ||
[[File:BMC 12-30.JPG|1024px]] | [[File:BMC 12-30.JPG|1024px]] | ||
(Pete Dunten had noticed that PDB code 2nls should be 1nls; this is now in the [http://www.ruppweb.org/garland/errata.htm book's errata]; the above legend has the correction.) | |||
---- | |||
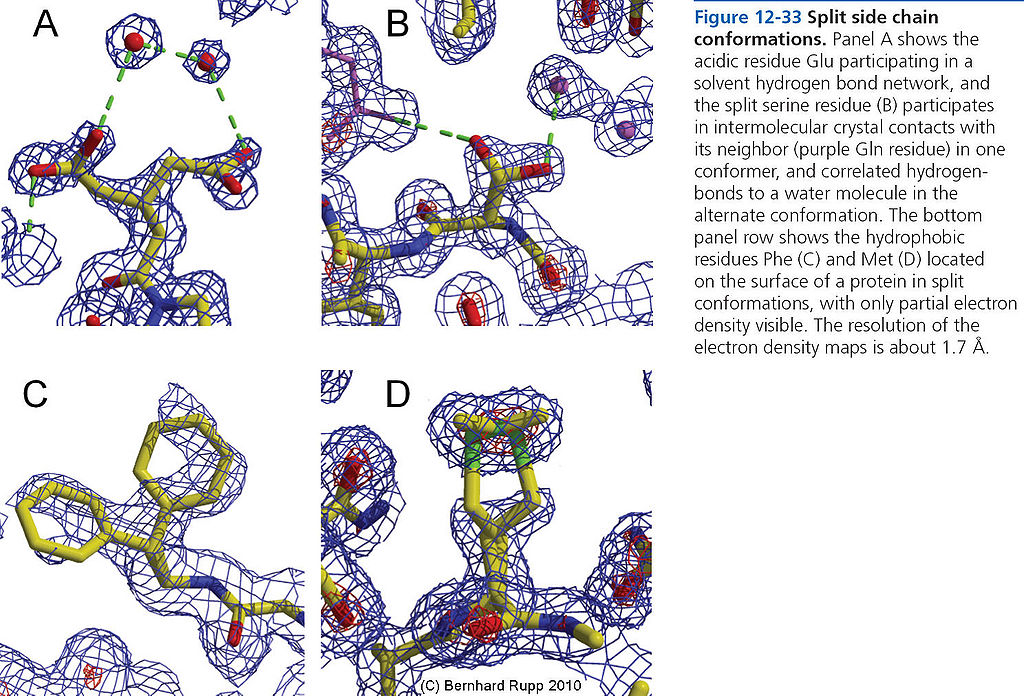
=== Split side-chain conformations === | === Split side-chain conformations === | ||
[[File:BMC 12-33.JPG|1024px]] | [[File:BMC 12-33.JPG|1024px]] | ||
---- | |||
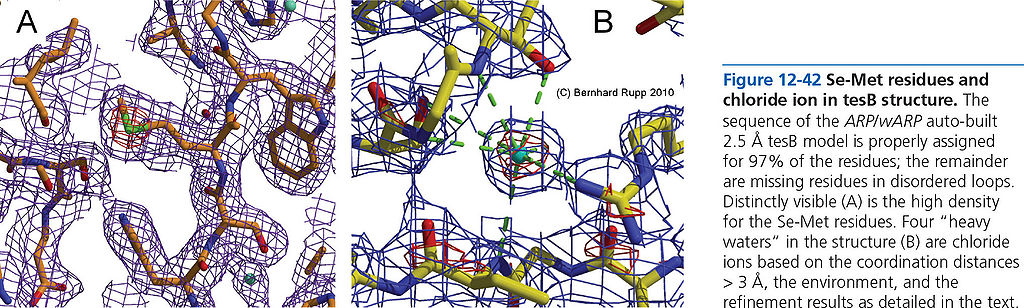
=== Se-Met residues and chloride ion === | === Se-Met residues and chloride ion === | ||
Latest revision as of 15:09, 19 July 2010
Figures from Bernhard Rupp's book "Biomolecular Crystallography" (with permission by the author)
There is more material (all figures, citations, tutorials) at the book's overview page.
You can see the full-size, high-resolution figure by clicking on it.
Cis-peptide bond and its electron density
Covalently modified residues
Covalent binding of ligands
Additives mediating intermolecular contacts
Heavy-atom derivatization of a protein
The trimeric bacterial outer membrane protein OprP
Chemically modified lysine residue (Dimethyllysine)
Glycosylated residues stabilizing homodimer
Uranium-atom solution and Fourier truncation ripples
Solvent molecules (water, MPD, Mg2+)
(Pete Dunten had noticed that PDB code 2nls should be 1nls; this is now in the book's errata; the above legend has the correction.)