1ZTV: Difference between revisions
Jump to navigation
Jump to search
| Line 1: | Line 1: | ||
== 1ZTV = Crystal structure of (29342463) from Enterococcus faecalis V583 at 3.10 A resolution == | == 1ZTV = Crystal structure of (29342463) from Enterococcus faecalis V583 at 3.10 A resolution == | ||
There are several datasets available from the JCSG dataset archive: | There are [http://www.jcsg.org/prod/scripts/data_repository/dataset_download.cgi?pdb_id=1ztv several datasets] available from the JCSG dataset archive (one needs an account for that!): | ||
native: | native: | ||
* 11258_1 : I/sigmaI for individual reflections (the fine-resolution list in CORRECT.LP) drops below 1 at 3.0 A resoluion. Space group is P212121 with a=91.91 b=92.50 c=119.60 A. This corresponds to the deposited structure. | * [[11258_1]] : I/sigmaI for individual reflections (the fine-resolution list in CORRECT.LP) drops below 1 at 3.0 A resoluion. Space group is P212121 with a=91.91 b=92.50 c=119.60 A. This corresponds to the deposited structure. | ||
MAD: | MAD: | ||
* 11859_1_E1, 11859_1_E2, 11859_1_E3: at wavelengths of 0.979694, 1.019859, 0.979571 (from the headers) these correspond to inflection, remote low and peak of a SeMet MAD experiment (these are guesses). This crystal is space group P65, with cell parameters a=b=114.28, c=52.4 A. | * [[11859_1_E1]], [[11859_1_E2]], [[11859_1_E3]]: at wavelengths of 0.979694, 1.019859, 0.979571 (from the headers) these correspond to inflection, remote low and peak of a SeMet MAD experiment (these are guesses). This crystal is space group P65, with cell parameters a=b=114.28, c=52.4 A. | ||
These data can be nicely analyzed using the hkl2map GUI. | These data can be nicely analyzed using the hkl2map GUI. | ||
Revision as of 13:50, 29 February 2008
1ZTV = Crystal structure of (29342463) from Enterococcus faecalis V583 at 3.10 A resolution
There are several datasets available from the JCSG dataset archive (one needs an account for that!):
native:
- 11258_1 : I/sigmaI for individual reflections (the fine-resolution list in CORRECT.LP) drops below 1 at 3.0 A resoluion. Space group is P212121 with a=91.91 b=92.50 c=119.60 A. This corresponds to the deposited structure.
MAD:
- 11859_1_E1, 11859_1_E2, 11859_1_E3: at wavelengths of 0.979694, 1.019859, 0.979571 (from the headers) these correspond to inflection, remote low and peak of a SeMet MAD experiment (these are guesses). This crystal is space group P65, with cell parameters a=b=114.28, c=52.4 A.
These data can be nicely analyzed using the hkl2map GUI.
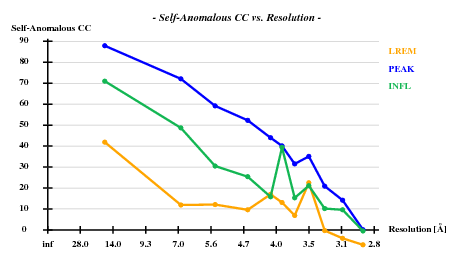
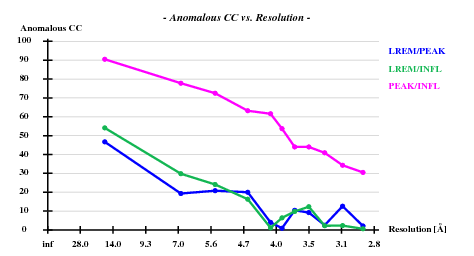
SHELXC (Versions 2006/3) reports
Correlation coefficients (%) between signed anomalous differences Resl. Inf - 8.0 - 6.0 - 5.0 - 4.2 - 4.0 - 3.8 - 3.6 - 3.4 - 3.2 - 3.0 - 2.80 LREM/PEAK 46.7 19.3 20.8 20.0 4.0 0.9 10.4 9.2 2.5 12.6 2.1 LREM/INFL 54.1 29.8 24.1 16.3 1.1 6.4 9.8 12.3 2.2 2.3 0.6 PEAK/INFL 90.5 77.8 72.5 63.2 61.6 53.7 44.0 44.0 40.9 34.3 30.5
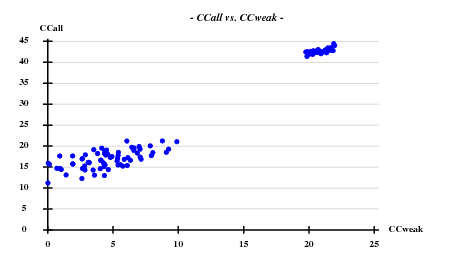
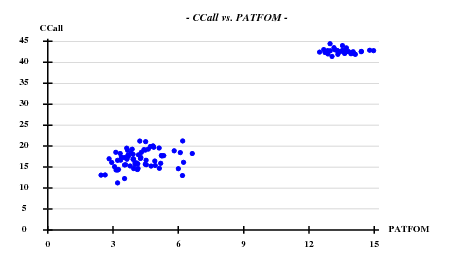
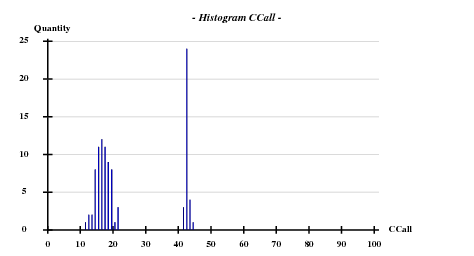
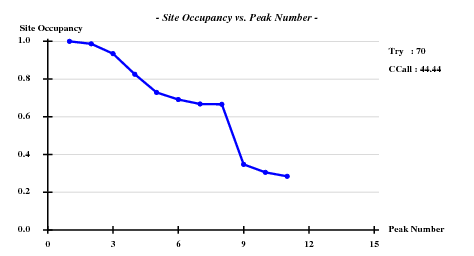
Searching 8 sites (after counting the Met in the sequence, and omitting the N-terminal one) with SHELXD-2006/3 with data to 3.3 A:
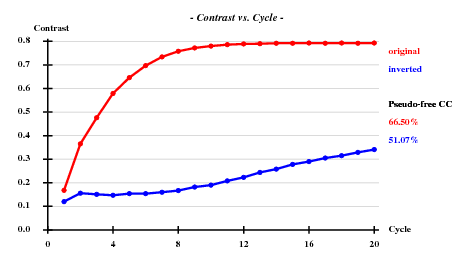
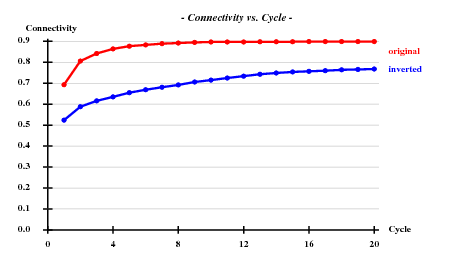
SHELXE (290 residues, solvent content calculated as ) suggests P65 instead of P61:
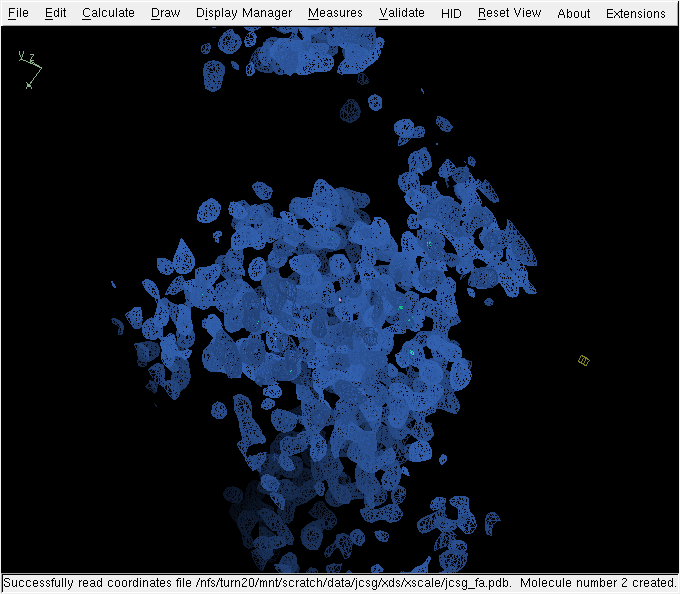
The structure can almost be considered solved, and (for this resolution) the density is quite nice:
References
- Pape T & Schneider TR (2004). HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Cryst. 37:843-844.
- SHELXC: Sheldrick GM (2003). Goettingen University.
- SHELXD: Schneider TR, Sheldrick GM (2002). Substructure solution with SHELXD. Acta Cryst. D58:1772-1779.
- SHELXE: Sheldrick GM (2002). Macromolecular phasing with SHELXE. Z. Kristallogr. 217:644-650.