1RQW: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
mNo edit summary |
||
| Line 1: | Line 1: | ||
==Thaumatin structure solution from either 2-wavelength MAD, or from | ==Thaumatin Br soak: structure solution from either 2-wavelength MAD, or from single wavelength SAD== | ||
Prior to flash cooling in liquid nitrogen at 100 K the crystals were soaked for a few seconds in a solution containing 1 M sodium bromide and 25% (v/v) glycerol. | |||
The peak and inflection datasets (360 images each) are available from the [http://www.embl-hamburg.de EMBL Hamburg] website. The XDS data reductions are [ftp://turn5.biologie.uni-konstanz.de//pub/xds-datared/1rqw here]. | The peak and inflection datasets (360 images each) are available from the [http://www.embl-hamburg.de EMBL Hamburg] website. The XDS data reductions are [ftp://turn5.biologie.uni-konstanz.de//pub/xds-datared/1rqw here]. | ||
| Line 32: | Line 34: | ||
=== automatic structure solution using Auto-Rickshaw === | === automatic structure solution using Auto-Rickshaw === | ||
DPS2AR.csh datafile1 thau-peak-I.mtz keeps world ver completeversion nres 207 nsm 20 masu 1 meth SAD SG P41212 email kay.diederichs@uni-konstanz.de sequencefile 1rqw.seq | |||
This uses Santosh Panjikar's script DPS2AR.csh: | |||
DPS2AR.csh datafile1 thau-peak-I.mtz keeps world ver completeversion nres 207 nsm 20 masu 1 meth SAD SG P41212 \ | |||
email kay.diederichs@uni-konstanz.de sequencefile 1rqw.seq | |||
where 1rqw.seq is | where 1rqw.seq is | ||
Revision as of 19:03, 30 April 2008
Thaumatin Br soak: structure solution from either 2-wavelength MAD, or from single wavelength SAD
Prior to flash cooling in liquid nitrogen at 100 K the crystals were soaked for a few seconds in a solution containing 1 M sodium bromide and 25% (v/v) glycerol.
The peak and inflection datasets (360 images each) are available from the EMBL Hamburg website. The XDS data reductions are here.
Data reduction was performed starting with the template for the MarCCD 225 detector, changing
ORGX=1533 ORGY=1536 DETECTOR_DISTANCE=200 OSCILLATION_RANGE=0.5 X-RAY_WAVELENGTH=0.91878 ! peak ! X-RAY_WAVELENGTH=0.9196 ! inflection NAME_TEMPLATE_OF_DATA_FRAMES=../peak/thau_peak_1_???.mccd DIRECT TIFF ! NAME_TEMPLATE_OF_DATA_FRAMES=../inf/thau_inf_1_???.mccd DIRECT TIFF DATA_RANGE=1 360 SPACE_GROUP_NUMBER=92 UNIT_CELL_CONSTANTS= 57.8 57.8 150. 90 90 90 FRIEDEL'S_LAW=FALSE VALUE_RANGE_FOR_TRUSTED_DETECTOR_PIXELS=9000. 30000.
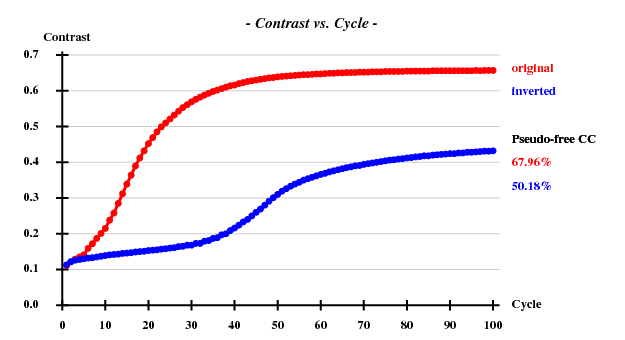
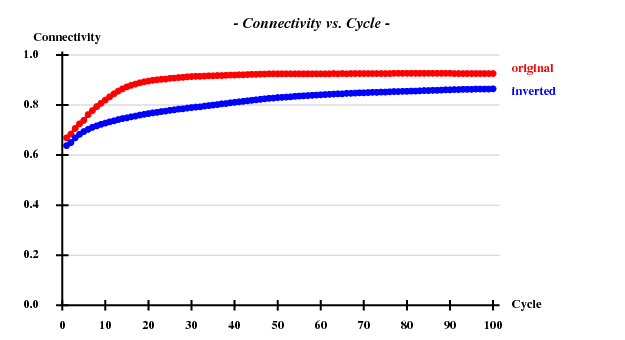
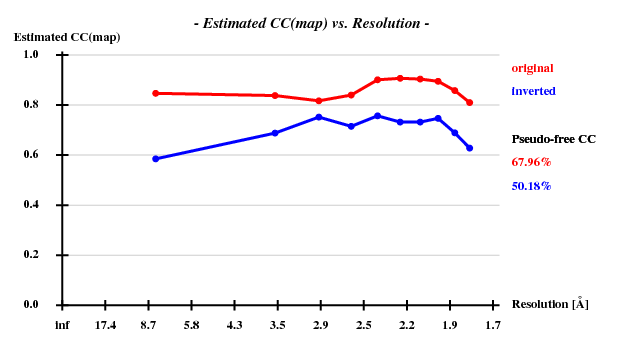
SAD: Peak data alone
manual structure solution using hkl2map and buccaneer
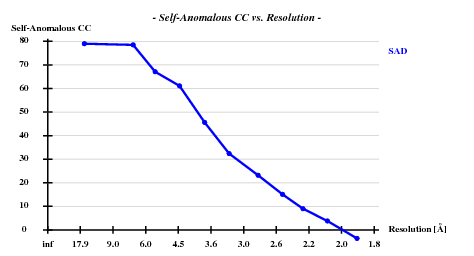
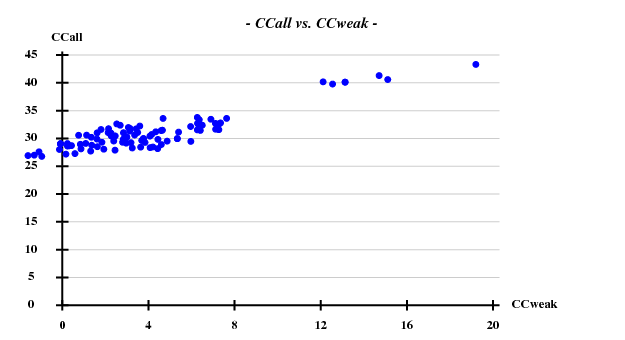
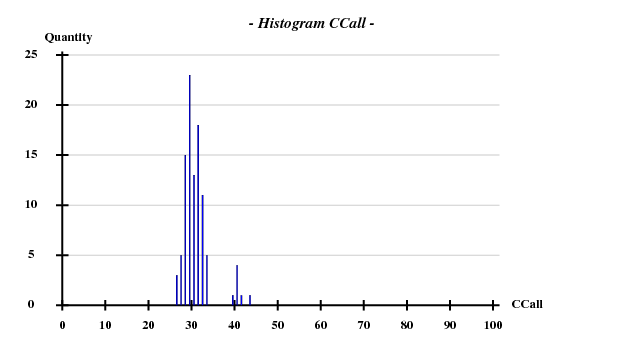
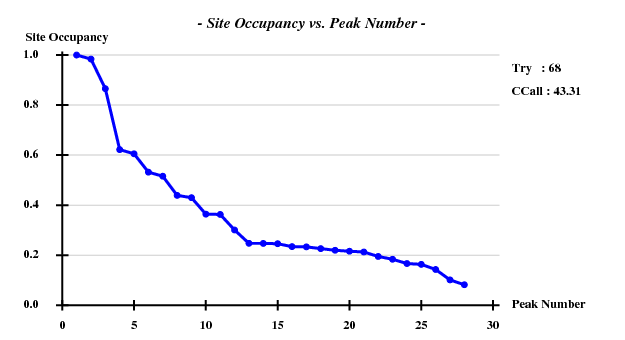
The structure was solved using the hkl2map GUI. Based on
I decided to use 3.3 Å as a suitable cutoff for solving the substructure. SHELXD then found
and SHELXE produced useful phases:
automatic structure solution using Auto-Rickshaw
This uses Santosh Panjikar's script DPS2AR.csh:
DPS2AR.csh datafile1 thau-peak-I.mtz keeps world ver completeversion nres 207 nsm 20 masu 1 meth SAD SG P41212 \
email kay.diederichs@uni-konstanz.de sequencefile 1rqw.seq
where 1rqw.seq is
ATFEIVNRCS YTVWAAASKG DAALDAGGRQ LNSGESWTIN VEPGTKGGKI WARTDCYFDD SGSGICKTGD CGGLLRCKRF GRPPTTLAEF SLNQYGKDYI DISNIKGFNV PMDFSPTTRG CRGVRCAADI VGQCPAKLKA PGGGCNDACT VFQTSEYCCT TGKCGPTEYS RFFKRLCPDA FSYVLDKPTT VTCPGSSNYR VTFCPTA