Space group determination: Difference between revisions
m (→Subgroup and supergroup relations of these space groups: add reference to ITC Vol A figure) |
|||
| (75 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
=== Space groups as combinations of symmetry elements === | |||
== Space group selected by | Space group determination entails the following steps: | ||
# determine the '''Laue class''': this is the symmetry of the intensity-weighted point lattice (diffraction pattern). 1,2,3,4,6=n-fold rotation axis; -n means inversion centre (normally the - is written over the n); m means mirror. | |||
# find out about '''Bravais type''': the second letter signifies centering (P=primitive; C=centered on C-face; F=centered on all faces; I=body-centered; R=rhombohedral). The first letter (a=triclinic; m=monoclinic; o=orthorhombic; h=trigonal or hexagonal; c=cubic) is redundant since it can be inferred from the Laue group. | |||
# possible spacegroups are now given in '''columns 3 and 4'''; CORRECT always suggests the one given in column 3 but this is no more likely than those in column 4. | |||
# choose according to screw axis (according to the table "REFLECTIONS OF TYPE H,0,0 0,K,0 0,0,L OR EXPECTED TO BE ABSENT (*)" in [[CORRECT.LP]]); this may result in two possibilities (enantiomorphs). | |||
# determine correct enantiomorph - this usually means that one tries to solve the structure in both spacegroups, and only one gives a sensible result (like helices that are right-handed, amino acids of the L type). | |||
== Table of space groups by Laue class and Bravais type == | |||
{| cellpadding="10" cellspacing="0" border="1" | |||
! Laue class | |||
! Bravais type | |||
! spacegroup <br> number <br> suggested by <br> CORRECT | |||
! other possibilities (with screw axes) | |||
! alternative indexing <br> possible? | |||
! choosing among all spacegroup possibilities | |||
|- | |||
| -1 || aP || 1 ||-|||| | |||
|- | |||
| 2/m || mP || 3 || 4 |||| screw axis extinctions let you decide | |||
|- | |||
| 2/m || mC || 5 || - |||| The [[pointless]] article discusses why I2 (mI) should not be used. | |||
|- | |||
| mmm || oP || 16 || 17, 18, 19 |||| screw axis extinctions let you decide. The [[pointless]] article discusses why CCP4's 1017 (P2<sub>1</sub>22), 2017 (P22<sub>1</sub>2), 2018 (P2<sub>1</sub>22<sub>1</sub>) , 3018 (P22<sub>1</sub>2<sub>1</sub>) are not needed and should not be used. | |||
|- | |||
| mmm || oC || 21 || 20 |||| screw axis extinctions let you decide | |||
|- | |||
| mmm || oF || 22 ||-|||| | |||
|- | |||
| mmm || oI || 23 || 24 |||| screw axis extinctions do '''not''' let you decide because the I centering results in h+k+l=2n and the screw axis extinction 00l=2n is just a special case of that. 23/24 do '''not''' form an enantiomorphic, but a ''special'' pair (ITC A §3.5, p. 46 in the 1995 edition). | |||
|- | |||
| 4/m || tP || 75 || 76, 77, 78 ||k,h,-l|| screw axis extinctions let you decide, except between 76/78 enantiomorphs | |||
|- | |||
| 4/m || tI || 79 || 80 ||k,h,-l|| screw axis extinctions let you decide | |||
|- | |||
| 4/mmm || tP || 89 || 90, 91, 92, 93, 94, 95, 96 |||| screw axis extinctions let you decide, except between 91/95 and 92/96 enantiomorphs | |||
|- | |||
| 4/mmm || tI || 97 || 98 |||| screw axis extinctions let you decide | |||
|- | |||
| -3 || hP || 143 || 144, 145 ||-h,-k,l; k,h,-l; -k,-h,-l|| screw axis extinctions let you decide, except between 144/145 enantiomorphs | |||
|- | |||
| -3 || hR || 146 || - ||k,h,-l, and obverse (-h+k+l=3n) / reverse (h-k+l=3n)|| | |||
|- | |||
| -3/m || hP || 149 || 151, 153 ||k,h,-l|| screw axis extinctions let you decide, except between 151/153 enantiomorphs. Note: the twofold goes along the diagonal between a and b. | |||
|- | |||
| -3/m || hP || 150 || 152, 154 ||-h,-k,l|| screw axis extinctions let you decide, except between 152/154 enantiomorphs. Note: compared to previous line, the twofold goes along a. | |||
|- | |||
| -3/m || hR || 155 || - ||obverse/reverse|| | |||
|- | |||
| 6/m || hP || 168 || 169, 170, 171, 172, 173 ||k,h,-l|| screw axis extinctions let you decide, except between 169/170 and 171/172 enantiomorphs | |||
|- | |||
| 6/mmm || hP || 177 || 178, 179, 180, 181, 182 |||| screw axis extinctions let you decide, except between 178/179 and 180/181 enantiomorphs | |||
|- | |||
| m-3 || cP || 195 || 198 ||k,h,-l|| screw axis extinctions let you decide | |||
|- | |||
| m-3 || cF || 196 ||-||k,h,-l|| | |||
|- | |||
| m-3 || cI || 197 || 199 ||k,h,-l|| screw axis extinctions do '''not''' let you decide because the I centering results in h+k+l=2n and the screw axis extinction 00l=2n is just a special case of that. 197/199 do '''not''' form an enantiomorphic, but a ''special'' pair (ITC A §3.5, p. 46 in the 1995 edition). | |||
|- | |||
| m-3m || cP || 207 || 208, 212, 213 |||| screw axis extinctions let you decide, except between 212/213 enantiomorphs | |||
|- | |||
| m-3m || cF || 209 || 210 |||| screw axis extinctions let you decide | |||
|- | |||
| m-3m || cI || 211 || 214 |||| screw axis extinctions let you decide | |||
|- | |||
|} | |||
Alternative indexing possibilities taken from http://www.ccp4.ac.uk/html/reindexing.html (better readable at http://www.csb.yale.edu/userguides/datamanip/ccp4/ccp4i/help/modules/appendices/reindexing.html) (for R3 and R32, obverse/reverse are specified). | |||
If you find an error in the table please send an email to kay dot diederichs at uni-konstanz dot de ! | |||
== The 65 Sohncke space groups in which proteins composed of L-amino acids can crystallize == | |||
The mapping of numbers and names is: | |||
****** LATTICE SYMMETRY IMPLICATED BY SPACE GROUP SYMMETRY ****** | |||
BRAVAIS- POSSIBLE SPACE-GROUPS FOR PROTEIN CRYSTALS | |||
TYPE [SPACE GROUP NUMBER,SYMBOL] | |||
aP [1,P1] | |||
mP [3,P2] [4,P2(1)] | |||
mC,mI [5,C2] | |||
oP [16,P222] [17,P222(1)] [18,P2(1)2(1)2] [19,P2(1)2(1)2(1)] | |||
oC [21,C222] [20,C222(1)] | |||
oF [22,F222] | |||
oI [23,I222] [24,I2(1)2(1)2(1)] | |||
tP [75,P4] [76,P4(1)] [77,P4(2)] [78,P4(3)] [89,P422] [90,P42(1)2] | |||
[91,P4(1)22] [92,P4(1)2(1)2] [93,P4(2)22] [94,P4(2)2(1)2] | |||
[95,P4(3)22] [96,P4(3)2(1)2] | |||
tI [79,I4] [80,I4(1)] [97,I422] [98,I4(1)22] | |||
hP [143,P3] [144,P3(1)] [145,P3(2)] [149,P312] [150,P321] [151,P3(1)12] | |||
[152,P3(1)21] [153,P3(2)12] [154,P3(2)21] [168,P6] [169,P6(1)] | |||
[170,P6(5)] [171,P6(2)] [172,P6(4)] [173,P6(3)] [177,P622] | |||
[178,P6(1)22] [179,P6(5)22] [180,P6(2)22] [181,P6(4)22] [182,P6(3)22] | |||
hR [146,R3] [155,R32] | |||
cP [195,P23] [198,P2(1)3] [207,P432] [208,P4(2)32] [212,P4(3)32] | |||
[213,P4(1)32] | |||
cF [196,F23] [209,F432] [210,F4(1)32] | |||
cI [197,I23] [199,I2(1)3] [211,I432] [214,I4(1)32] | |||
=== Subgroup and supergroup relations of these space groups === | |||
Compiled from [https://onlinelibrary.wiley.com/doi/book/10.1107/97809553602060000001 International Tables for Crystallography (2006) Vol. A1 (Wiley)]. Simply put, for each space group, a maximum ''translationengleiche'' subgroup has lost a single type of symmetry, and a minimum ''translationengleiche'' supergroup has gained a single symmetry type. Example: P222 is a supergroup of P2, and a subgroup of P422 (and P4222 and P23). Of course the sub-/supergroup relation is recursive, which is why P1 is also a (sub-)subgroup of P222 (but not a maximum ''translationengleiche'' subgroup). The table below does not show other types of relations, e.g. non-isomorphic ''klassengleiche'' supergroups which may result e.g. from centring translations, because I find them less relevant in space group determination. | |||
The table is relevant because in particular (perfect) twinning adds a symmetry type, and leads to an apparent space group which is the supergroup of the true space group. | |||
{| cellpadding="0" cellspacing="0" border="1" | |||
! spacegroup number | |||
! maximum ''translationengleiche'' subgroup | |||
! minimum ''translationengleiche'' supergroup | |||
! spacegroup name | |||
|- | |||
| 1 ||-|| 3, 4, 5, 143, 144, 145, 146 || P 1 | |||
|- | |||
| 3 || 1 || 16, 17, 18, 21, 75, 77, 168, 171, 172 || P 2 | |||
|- | |||
| 4 || 1 || 17, 18, 19, 20, 76, 78, 169, 170, 173 || P 21 | |||
|- | |||
| 5 || 1 || 20, 21, 22, 23, 24, 79, 80, 149, 150, 151, 152, 153, 154, 155 || C2 | |||
|- | |||
| 16 || 3 || 89, 93, 195 || P 2 2 2 | |||
|- | |||
| 17 || 3, 4 || 91, 95 || P 2 2 21 | |||
|- | |||
| 18 || 3, 4 || 90, 94 || P 21 21 2 | |||
|- | |||
| 19 || 4 || 92, 96, 198 || P 21 21 21 | |||
|- | |||
| 20 || 4, 5 || 91, 92, 95, 96, 178, 179, 182 || C 2 2 21 | |||
|- | |||
| 21 || 3, 5 || 89, 90, 93, 94, 177, 180, 181 || C 2 2 2 | |||
|- | |||
| 22 || 5 || 97, 98, 196 || F 2 2 2 | |||
|- | |||
| 23 || 5 || 97, 197 || I 2 2 2 | |||
|- | |||
| 24 || 5 || 98, 199 || I 21 21 21 | |||
|- | |||
| 75 || 3 || 89, 90 || P 4 | |||
|- | |||
| 76 || 4 || 91, 92 || P 41 | |||
|- | |||
| 77 || 3 || 93, 94 || P 42 | |||
|- | |||
| 78 || 4 || 95, 96 || P 43 | |||
|- | |||
| 79 || 5 || 97 || I 4 | |||
|- | |||
| 80 || 5 || 98 || I 41 | |||
|- | |||
| 89 || 16, 21, 75 || 207 || P 4 2 2 | |||
|- | |||
| 90 || 18, 21, 75 || - || P 4 21 2 | |||
|- | |||
| 91 || 17, 20, 76 || - || P 41 2 2 | |||
|- | |||
| 92 || 19, 20, 76 || 213 || P 41 21 2 | |||
|- | |||
| 93 || 16, 21, 77 || 208 || P 42 2 2 | |||
|- | |||
| 94 || 18, 21, 77 || 93, 97 || P 42 21 2 | |||
|- | |||
| 95 || 17, 20, 78 || - || P 43 2 2 | |||
|- | |||
| 96 || 19, 20, 78 || 212 || P 43 21 2 | |||
|- | |||
| 97 || 22, 23, 79 || 209, 211 || I 4 2 2 | |||
|- | |||
| 98 || 22, 24, 80 || 210, 214 || I 41 2 2 | |||
|- | |||
| 143 || 1 || 149, 150, 168, 173 || P 3 | |||
|- | |||
| 144 || 1 || 151, 152, 169, 172 || P 31 | |||
|- | |||
| 145 || 1 || 153, 154, 170, 171 || P 32 | |||
|- | |||
| 146 || 1 || 155, 195, 196, 197, 198, 199 || R 3 | |||
|- | |||
| 149 || 5, 143 || 177, 182 || P 3 1 2 | |||
|- | |||
| 150 || 5, 143 || 177, 182 || P 3 2 1 | |||
|- | |||
| 151 || 5, 144 || 178, 181 || P 31 1 2 | |||
|- | |||
| 152 || 5, 144 || 178, 181 || P 31 2 1 | |||
|- | |||
| 153 || 5, 145 || 179, 180 || P 32 1 2 | |||
|- | |||
| 154 || 5, 145 || 179, 180 || P 32 2 1 | |||
|- | |||
| 155 || 5, 146 || 207, 208, 209, 210, 211, 212, 213, 214 || R 3 2 | |||
|- | |||
| 168 || 3, 143 || 177 || P 6 | |||
|- | |||
| 169 || 4, 144 || 178 || P 61 | |||
|- | |||
| 170 || 4, 145 || 179 || P 65 | |||
|- | |||
| 171 || 3, 145 || 180 || P 62 | |||
|- | |||
| 172 || 3, 144 || 181 || P 64 | |||
|- | |||
| 173 || 4, 143 || 182 || P 63 | |||
|- | |||
| 177 || 21, 149, 150, 168 || - || P 6 2 2 | |||
|- | |||
| 178 || 20, 151, 152, 169 || - || P 61 2 2 | |||
|- | |||
| 179 || 20, 153, 154, 170 || - || P 65 2 2 | |||
|- | |||
| 180 || 21, 153, 154, 171 || - || P 62 2 2 | |||
|- | |||
| 181 || 21, 151, 152, 172 || - || P 64 2 2 | |||
|- | |||
| 182 || 20, 149, 150, 173 || - || P 63 2 2 | |||
|- | |||
| 195 || 16, 146 || 207, 208 || P 2 3 | |||
|- | |||
| 196 || 22, 146 || 209, 210 || F 2 3 | |||
|- | |||
| 197 || 23, 146 || 211 || I 2 3 | |||
|- | |||
| 198 || 19, 146 || 212, 213 || P 21 3 | |||
|- | |||
| 199 || 24, 146 || 214 || I 21 3 | |||
|- | |||
| 207 || 89, 155, 195 || - || P 4 3 2 | |||
|- | |||
| 208 || 93, 155, 195 || - || P 42 3 2 | |||
|- | |||
| 209 || 97, 155, 196 || - || F 4 3 2 | |||
|- | |||
| 210 || 98, 155, 196 || - || F 41 3 2 | |||
|- | |||
| 211 || 97, 155, 197 || - || I 4 3 2 | |||
|- | |||
| 212 || 96, 155, 198 || - || P 43 3 2 | |||
|- | |||
| 213 || 92, 155, 198 || - || P 41 3 2 | |||
|- | |||
| 214 || 98, 155, 199 || - || I 41 3 2 | |||
|- | |||
|} | |||
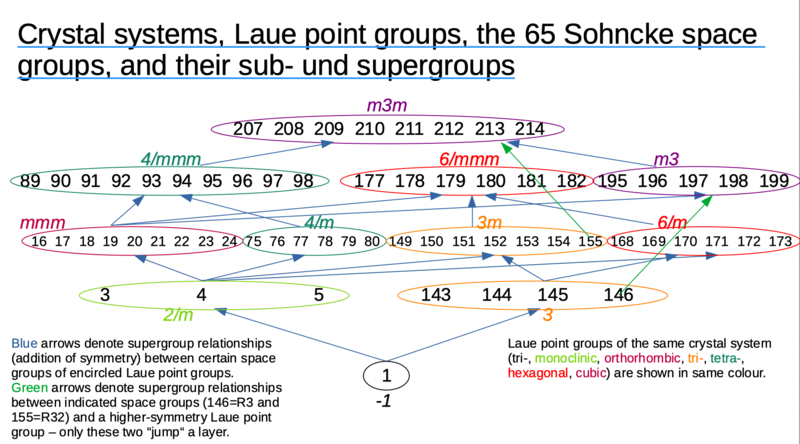
[[File:Spacegroups tree2a.png|800px]] | |||
(compare International Tables for Crystallography Vol A (2006), figure 10.1.3.2) | |||
== Space group selected by XDS: ambiguous with respect to enantiomorph and screw axes == | |||
In case of a crystal with an unknown space group (SPACE_GROUP_NUMBER=0 in [[XDS.INP]]), XDS (since [http://xds.mpimf-heidelberg.mpg.de/html_doc/Release_Notes.html version June 2008]) helps the user in determination of the correct space group, by suggesting possible space groups compatible with the Laue symmetry and Bravais type of the data, and by calculating the R<sub>meas</sub> for these space groups. | |||
XDS (or rather, the [[CORRECT.LP|CORRECT]] step) makes an attempt to pick the correct space group automatically: it chooses the space group (or rather: Laue point group) which has the highest symmetry (thus yielding the lowest number of unique reflections) and still a tolerable R<sub>meas</sub> compared to the R<sub>meas</sub> the data have in any space group (which is most likely a low-symmetry space group - often P1). | |||
In some cases the automatic choice is the correct one, and re-running the [[CORRECT.LP|CORRECT]] step is then not necessary. However, neither the correct enantiomorph nor [[Space_group_determination#Screw_axes|screw axes]] (see below) are determined automatically by XDS. [[Pointless]] is a very good program (usually better than CORRECT) to suggest possible space group (and alternatives). See also [[Space_group_determination#Notes|Notes]], and [[Space_group_determination#checking_the_CORRECT_assignment_with_pointless|checking the CORRECT assignment with pointless]] (below). | |||
== Space group selected by user == | |||
=== | In case the space group selected by XDS should be incorrect, the resulting list (in CORRECT.LP) should give the user enough information to pick the correct space group herself (alternatives are in the big table above!). The user may then put suitable lines with SPACE_GROUP_NUMBER=, UNIT_CELL_CONSTANTS= into XDS.INP and re-run the CORRECT step to obtain the desired result. (The REIDX= line is no longer required; XDS figures the matrix out.) | ||
== Influencing the selection by XDS == | |||
=== keywords and parameters === | |||
The automatic choice is influenced by a number of decision constants that may be put into XDS.INP but which have defaults as indicated below: | |||
* MAX_CELL_AXIS_ERROR= 0.03 ! relative deviation of unconstrained cell axes from those constrained by lattice symmetry | * MAX_CELL_AXIS_ERROR= 0.03 ! relative deviation of unconstrained cell axes from those constrained by lattice symmetry | ||
* MAX_CELL_ANGLE_ERROR= 0 | * MAX_CELL_ANGLE_ERROR= 3.0 ! degrees deviation of unconstrained cell angles from those constrained by lattice symmetry | ||
* TEST_RESOLUTION_RANGE= 10.0 5.0 ! resolution range for calculation of R<sub>meas</sub> | * TEST_RESOLUTION_RANGE= 10.0 5.0 ! resolution range for calculation of R<sub>meas</sub> | ||
* MIN_RFL_Rmeas= 50 ! at least this number of reflections are required | * MIN_RFL_Rmeas= 50 ! at least this number of reflections are required | ||
* MAX_FAC_Rmeas= 2.0 ! factor to multiply | * MAX_FAC_Rmeas= 2.0 ! factor to multiply the lowest R<sub>meas</sub> with to still be acceptable | ||
The user may experiment with adjusting these values to make the automatic mode of space group determination more successful. For example, if the crystal diffracts weakly, all R<sub>meas</sub> values will be high and no valid decision can be made. In this case, I suggest to use e.g. | |||
TEST_RESOLUTION_RANGE= 50.0 10.0 | |||
=== dealing with alternative indexing === | |||
There are two ways to have XDS choose an indexing consistent with some other dataset: | |||
* using REFERENCE_DATA_SET= ! see also [[REFERENCE_DATA_SET]] | |||
* using [http://xds.mpimf-heidelberg.mpg.de/html_doc/xds_parameters.html#UNIT_CELL_A-AXIS= UNIT_CELL_A-AXIS=], UNIT_CELL_B-AXIS=, UNIT_CELL_C-AXIS= from a previous data collection run with the same crystal | |||
One can also manually force a specific indexing, using the REIDX= keyword, but this is error-prone. | |||
See also http://www.ccp4.ac.uk/html/reindexing.html | |||
== Screw axes == | |||
The user | The current version makes no attempt to find out about screw axes. It is assumed that the user checks the table in CORRECT.LP entitled '''REFLECTIONS OF TYPE H,0,0 0,K,0 0,0,L OR EXPECTED TO BE ABSENT (*)''', and identifies whether the intensities follow the rules | ||
# a two-fold screw axis along an axis in reciprocal space (theoretically) results in zero intensity for the odd-numbered (e.g. 0,K,0 with K = 2*n + 1) reflections, leaving the reflections of type 2*n as candidates for medium to strong reflections (they don't ''have'' to be strong, but they ''may'' be strong!). | |||
# similarly, a three-fold screw axis (theoretically) results in zero intensity for the reflections of type 3*n+1 and 3*n+2, and allows possibly strong 3*n reflections . 3<sub>1</sub> and 3<sub>2</sub> cannot be distinguished - they are enantiomorphs. | |||
# analogously for four-fold screw axes: for H=0, K=0 reflections, 4<sub>1</sub> and 4<sub>3</sub> screws yield the rule L=4*n, and 4<sub>2</sub> yields the rule L=2*n . | |||
# analogously for six-fold screw axes: for H=0, K=0 reflections, 6<sub>1</sub> and 6<sub>5</sub> screws yield the rule L=6*n, 6<sub>2</sub> and 6<sub>4</sub> yield the rule L=3*n, and 6<sub>3</sub> yields the rule L=2*n . | |||
Once screw axes have been deduced from the patterns of intensities along H,0,0 0,K,0 0,0,L , the resulting space group should be identified in the lists of space group numbers printed in IDXREF.LP and CORRECT.LP, and the CORRECT step can be re-run. Those reflections that should theoretically have zero intensity are then marked with a "*" in the table. In practice, they should be (hopefully quite) weak, or even negative. | |||
== Notes == | == Notes == | ||
There is still the page [[Old way of Space group determination]] | * There is still the old Wiki page [[Old way of Space group determination]] | ||
* To prevent XDS from trying to find a better space group than the one with the lowest R<sub>meas</sub> (often P1), you could just use MAX_FAC_Rmeas= 1.0 | |||
* Space group determination is not a trivial task. There is a number of difficulties that arise - | |||
** ambiguity of hand of screw axis (e.g. 3<sub>1</sub> versus 3<sub>2</sub>, or 6<sub>2</sub> versus 6<sub>4</sub>) - see the table above! | |||
** twinning may make a low-symmetry space group look like a high-symmetry one | |||
* [[pointless]] helps with identification of screw axes, and its identification of Laue group is more sensitive than that implemented in CORRECT. However it is not fail-safe, and cannot tell the correct enantiomorph. | |||
* only when the structure is satisfactorily refined can the chosen space group be considered established and correct. Until then, it is just a hypothesis and its alternatives (see the table) should be kept in mind. | |||
== An example == | |||
The following example shows the difficulty in deciding between higher and lower symmetry. | |||
IDXREF reports: | |||
<pre> | |||
LATTICE- BRAVAIS- QUALITY UNIT CELL CONSTANTS (ANGSTROEM & DEGREES) | |||
CHARACTER LATTICE OF FIT a b c alpha beta gamma | |||
* 44 aP 0.0 74.0 78.6 124.0 108.2 105.1 90.4 | |||
* 31 aP 3.5 74.0 78.6 124.0 71.8 74.9 90.4 | |||
* 41 mC 7.4 235.7 78.6 74.0 90.4 106.1 89.7 | |||
* 37 mC 34.8 239.6 74.0 78.6 90.4 109.0 87.8 | |||
* 42 oI 38.7 74.0 78.6 226.6 90.2 92.2 90.4 | |||
14 mC 122.5 107.6 108.3 124.0 92.8 114.1 86.6 | |||
... | |||
</pre> | |||
so oI (space groups 23=I222 or 24=I2<sub>1</sub>2<sub>1</sub>2<sub>1</sub>) or the two mC (space group 5=C2) are possibilities for indexing, and of course P1. Integration in P1 (because of SPACE_GROUP_NUMBER=0 in XDS.INP) results in the following table for space group determination in CORRECT: | |||
<pre> | |||
SPACE-GROUP UNIT CELL CONSTANTS UNIQUE Rmeas COMPARED LATTICE- | |||
NUMBER a b c alpha beta gamma CHARACTER | |||
1 74.1 78.6 124.0 71.8 74.8 90.4 6959 22.5 5309 31 aP | |||
* 23 74.1 78.6 226.6 90.0 90.0 90.0 2735 39.5 9533 42 oI | |||
5 235.7 78.6 74.1 90.0 106.1 90.0 3922 31.0 8346 41 mC | |||
5 239.6 74.1 78.6 90.0 109.0 90.0 5111 30.7 7157 37 mC | |||
1 74.1 78.6 124.0 108.2 105.2 90.4 6959 22.5 5309 44 aP | |||
</pre> | |||
The automatic choice gave 23 as a result. This is because the associated R<sub>meas</sub> (39.5) is lower than MAX_FAC_RMEAS * (lowest R<sub>meas</sub>) = 2 * 22.5 = 45.0. | |||
However, the C2 values of 30.7 and 31.0 are significantly lower still, and the question needs further investigation. | |||
=== checking the CORRECT assignment with [[pointless]] === | |||
<code>[[pointless]] XDS_ASCII.HKL</code> gives: | |||
<pre> | |||
Analysing rotational symmetry in lattice group I 4/m m m | |||
---------------------------------------------- | |||
<!--SUMMARY_BEGIN--> | |||
Scores for each symmetry element | |||
Nelmt Lklhd Z-cc CC N Rmeas Symmetry & operator (in Lattice Cell) | |||
1 0.856 7.77 0.78 9368 0.224 identity | |||
2 0.139 2.98 0.30 6376 0.894 2-fold l ( 0 0 1) {-h,-k,l} | |||
3 0.842 8.02 0.80 18679 0.234 ** 2-fold k ( 0 1 0) {-h,k,-l} | |||
4 0.148 3.10 0.31 5858 1.254 2-fold h ( 1 0 0) {h,-k,-l} | |||
5 0.073 1.25 0.13 12111 1.077 2-fold ( 1-1 0) {-k,-h,-l} | |||
6 0.071 1.11 0.11 12293 1.152 2-fold ( 1 1 0) {k,h,-l} | |||
7 0.068 0.83 0.08 24758 1.084 4-fold l ( 0 0 1) {-k,h,l}{k,-h,l} | |||
</pre> | |||
and this shows that actually there is only one instead of three two-fold axes, despite the fact that CORRECT auto-chose the orthorhombic system and possibly even rejected some reflections that do not fit it well. | |||
Pointless then re-indexes to I2 and suggests: | |||
<pre> | |||
Best Solution: space group I 1 2 1 | |||
Reindex operator: [h,k,l] | |||
Laue group probability: 0.763 | |||
Systematic absence probability: 1.000 | |||
Total probability: 0.763 | |||
Space group confidence: 0.687 | |||
Laue group confidence 0.687 | |||
Unit cell: 78.05 80.78 235.63 90.00 90.00 90.00 | |||
50.67 to 3.73 - Resolution range used for Laue group search | |||
50.67 to 3.08 - Resolution range in file, used for systematic absence check | |||
Number of batches in file: 120 | |||
The data do not appear to be twinned, from the L-test | |||
$$ <!--SUMMARY_END--> | |||
HKLIN spacegroup: I 2 2 2 body-centred orthorhombic | |||
$TEXT:Warning:$$ $$ | |||
The input crystal system is body-centred orthorhombic | |||
(Cell: 78.05 80.78 235.63 90.00 90.00 90.00) | |||
The crystal system chosen for output is body-centred monoclinic | |||
(Cell: 78.05 80.78 235.63 90.00 90.00 90.00) | |||
$TEXT:Warning:$$ $$ | |||
WARNING: | |||
WARNING: | |||
The chosen output crystal system is different from that used for integration of the input file(s). | |||
You should rerun the integration in the chosen crystal system because the cell constraints differ | |||
$$ | |||
Filename: | |||
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ | |||
Final point group choice has alternative indexing possibilities | |||
Alternative indexing possibilities are marked '*' if the cells are | |||
too different at the maximum resolution | |||
CellDifference(A) ReindexOperator | |||
1 0.0 [h,k,l] | |||
2 0.0 [h,-k,-l] | |||
3 8.0 * [k,-h,l] | |||
4 8.0 * [-k,h,l] | |||
</pre> | |||
At the bottom of the output, we see that other indexing possibilities exist. That means that one has to be very careful when merging crystals from this project, and that there may also be the possibility of twinning. | |||
In this particular case, CORRECT chose the wrong symmetry, whereas pointless identified the correct symmetry elements. I have also seen cases where pointless mis-identified the symmetry, usually on the side of too high symmetry. To avoid mistakes in space group identification, it is absolutely crucial to read and understand the tables that the programs print. | |||
=== a more sensible way to run pointless === | |||
pointless should really be run with the "SETTING SYMMETRY-BASED" option. When doing that, the output changes to | |||
<pre> | |||
Best Solution: space group C 1 2 1 | |||
Reindex operator: [h+l,k,-h] | |||
Laue group probability: 0.807 | |||
Systematic absence probability: 1.000 | |||
Total probability: 0.807 | |||
Space group confidence: 0.734 | |||
Laue group confidence 0.734 | |||
Unit cell: 248.53 80.86 78.08 90.00 108.31 90.00 | |||
40.76 to 3.80 - Resolution range used for Laue group search | |||
40.76 to 3.08 - Resolution range in file, used for systematic absence check | |||
Number of batches in file: 120 | |||
The data do not appear to be twinned, from the L-test | |||
$$ <!--SUMMARY_END--> | |||
HKLIN spacegroup: I 2 2 2 body-centred orthorhombic | |||
$TEXT:Warning:$$ $$ | |||
The input crystal system is body-centred orthorhombic | |||
(Cell: 78.08 80.86 235.95 90.00 90.00 90.00) | |||
The crystal system chosen for output is C-centred monoclinic | |||
(Cell: 248.53 80.86 78.08 90.00 108.31 90.00) | |||
$TEXT:Warning:$$ $$ | |||
WARNING: | |||
WARNING: | |||
The chosen output crystal system is different from that used for integration of the input file(s). | |||
You should rerun the integration in the chosen crystal system because the cell constraints differ | |||
$$ | |||
</pre> | |||
so the spacegroup given is the more normal C2 setting (instead of A2 or I2). Unfortunately, pointless does not seem to print out the table of "alternative indexing possibilities" in this mode - but possibly the table is only printed in the I2 case because the beta angle comes out as 90.0°. | |||
=== not biasing the cell parameters, and avoiding premature outlier rejection === | |||
CORRECT can easily be forced ''not'' to assign a spacegroup, and consequently will not reject outliers based on a too high symmetry assignment. To this end one simply supplies space group P1 and its unit cell: | |||
<pre> | |||
SPACE_GROUP_NUMBER=1 | |||
UNIT_CELL_CONSTANTS= 74.1 78.6 124.0 108.2 105.2 90.4 | |||
</pre> | |||
in XDS.INP and re-runs CORRECT. That gives an unbiased XDS_ASCII.HKL, with no angles set to 90°. pointless ''with'' SETTING SYMMETRY_BASED then gives | |||
<pre> | |||
Best Solution: space group C 1 2 1 | |||
Reindex operator: [-k-2l,-k,-h] | |||
Laue group probability: 0.849 | |||
Systematic absence probability: 1.000 | |||
Total probability: 0.849 | |||
Space group confidence: 0.770 | |||
Laue group confidence 0.770 | |||
Unit cell: 233.70 78.42 73.22 90.31 105.34 89.77 | |||
</pre> | |||
and ''without'' SETTING SYMMETRY_BASED | |||
<pre> | |||
Best Solution: space group I 1 2 1 | |||
Reindex operator: [-h,-k,h+k+2l] | |||
Laue group probability: 0.838 | |||
Systematic absence probability: 1.000 | |||
Total probability: 0.838 | |||
Space group confidence: 0.753 | |||
Laue group confidence 0.753 | |||
Unit cell: 233.70 78.42 73.22 90.31 105.34 89.77 | |||
</pre> | |||
the dreadful I2 space group nomenclature (and I have no idea why the probability and confidence values are worse than in C2). But anyway this shows that there are two non-equivalent ways to index the same lattice! | |||
=== final steps === | |||
Finally, XDS (but only JOB=DEFPIX INTEGRATE CORRECT) should be re-run with | |||
<pre> | |||
SPACE_GROUP_NUMBER=5 | |||
UNIT_CELL_CONSTANTS= 233.70 78.42 73.22 90 105.34 90 | |||
</pre> | |||
because this enforces just the correct cell constraints. | |||
== See also == | |||
[http://pd.chem.ucl.ac.uk/pdnn/symm3/allsgp.htm The 230 3-Dimensional Space Groups] | |||
Latest revision as of 07:41, 6 September 2023
Space groups as combinations of symmetry elements
Space group determination entails the following steps:
- determine the Laue class: this is the symmetry of the intensity-weighted point lattice (diffraction pattern). 1,2,3,4,6=n-fold rotation axis; -n means inversion centre (normally the - is written over the n); m means mirror.
- find out about Bravais type: the second letter signifies centering (P=primitive; C=centered on C-face; F=centered on all faces; I=body-centered; R=rhombohedral). The first letter (a=triclinic; m=monoclinic; o=orthorhombic; h=trigonal or hexagonal; c=cubic) is redundant since it can be inferred from the Laue group.
- possible spacegroups are now given in columns 3 and 4; CORRECT always suggests the one given in column 3 but this is no more likely than those in column 4.
- choose according to screw axis (according to the table "REFLECTIONS OF TYPE H,0,0 0,K,0 0,0,L OR EXPECTED TO BE ABSENT (*)" in CORRECT.LP); this may result in two possibilities (enantiomorphs).
- determine correct enantiomorph - this usually means that one tries to solve the structure in both spacegroups, and only one gives a sensible result (like helices that are right-handed, amino acids of the L type).
Table of space groups by Laue class and Bravais type
| Laue class | Bravais type | spacegroup number suggested by CORRECT |
other possibilities (with screw axes) | alternative indexing possible? |
choosing among all spacegroup possibilities |
|---|---|---|---|---|---|
| -1 | aP | 1 | - | ||
| 2/m | mP | 3 | 4 | screw axis extinctions let you decide | |
| 2/m | mC | 5 | - | The pointless article discusses why I2 (mI) should not be used. | |
| mmm | oP | 16 | 17, 18, 19 | screw axis extinctions let you decide. The pointless article discusses why CCP4's 1017 (P2122), 2017 (P2212), 2018 (P21221) , 3018 (P22121) are not needed and should not be used. | |
| mmm | oC | 21 | 20 | screw axis extinctions let you decide | |
| mmm | oF | 22 | - | ||
| mmm | oI | 23 | 24 | screw axis extinctions do not let you decide because the I centering results in h+k+l=2n and the screw axis extinction 00l=2n is just a special case of that. 23/24 do not form an enantiomorphic, but a special pair (ITC A §3.5, p. 46 in the 1995 edition). | |
| 4/m | tP | 75 | 76, 77, 78 | k,h,-l | screw axis extinctions let you decide, except between 76/78 enantiomorphs |
| 4/m | tI | 79 | 80 | k,h,-l | screw axis extinctions let you decide |
| 4/mmm | tP | 89 | 90, 91, 92, 93, 94, 95, 96 | screw axis extinctions let you decide, except between 91/95 and 92/96 enantiomorphs | |
| 4/mmm | tI | 97 | 98 | screw axis extinctions let you decide | |
| -3 | hP | 143 | 144, 145 | -h,-k,l; k,h,-l; -k,-h,-l | screw axis extinctions let you decide, except between 144/145 enantiomorphs |
| -3 | hR | 146 | - | k,h,-l, and obverse (-h+k+l=3n) / reverse (h-k+l=3n) | |
| -3/m | hP | 149 | 151, 153 | k,h,-l | screw axis extinctions let you decide, except between 151/153 enantiomorphs. Note: the twofold goes along the diagonal between a and b. |
| -3/m | hP | 150 | 152, 154 | -h,-k,l | screw axis extinctions let you decide, except between 152/154 enantiomorphs. Note: compared to previous line, the twofold goes along a. |
| -3/m | hR | 155 | - | obverse/reverse | |
| 6/m | hP | 168 | 169, 170, 171, 172, 173 | k,h,-l | screw axis extinctions let you decide, except between 169/170 and 171/172 enantiomorphs |
| 6/mmm | hP | 177 | 178, 179, 180, 181, 182 | screw axis extinctions let you decide, except between 178/179 and 180/181 enantiomorphs | |
| m-3 | cP | 195 | 198 | k,h,-l | screw axis extinctions let you decide |
| m-3 | cF | 196 | - | k,h,-l | |
| m-3 | cI | 197 | 199 | k,h,-l | screw axis extinctions do not let you decide because the I centering results in h+k+l=2n and the screw axis extinction 00l=2n is just a special case of that. 197/199 do not form an enantiomorphic, but a special pair (ITC A §3.5, p. 46 in the 1995 edition). |
| m-3m | cP | 207 | 208, 212, 213 | screw axis extinctions let you decide, except between 212/213 enantiomorphs | |
| m-3m | cF | 209 | 210 | screw axis extinctions let you decide | |
| m-3m | cI | 211 | 214 | screw axis extinctions let you decide |
Alternative indexing possibilities taken from http://www.ccp4.ac.uk/html/reindexing.html (better readable at http://www.csb.yale.edu/userguides/datamanip/ccp4/ccp4i/help/modules/appendices/reindexing.html) (for R3 and R32, obverse/reverse are specified). If you find an error in the table please send an email to kay dot diederichs at uni-konstanz dot de !
The 65 Sohncke space groups in which proteins composed of L-amino acids can crystallize
The mapping of numbers and names is:
****** LATTICE SYMMETRY IMPLICATED BY SPACE GROUP SYMMETRY ******
BRAVAIS- POSSIBLE SPACE-GROUPS FOR PROTEIN CRYSTALS
TYPE [SPACE GROUP NUMBER,SYMBOL]
aP [1,P1]
mP [3,P2] [4,P2(1)]
mC,mI [5,C2]
oP [16,P222] [17,P222(1)] [18,P2(1)2(1)2] [19,P2(1)2(1)2(1)]
oC [21,C222] [20,C222(1)]
oF [22,F222]
oI [23,I222] [24,I2(1)2(1)2(1)]
tP [75,P4] [76,P4(1)] [77,P4(2)] [78,P4(3)] [89,P422] [90,P42(1)2]
[91,P4(1)22] [92,P4(1)2(1)2] [93,P4(2)22] [94,P4(2)2(1)2]
[95,P4(3)22] [96,P4(3)2(1)2]
tI [79,I4] [80,I4(1)] [97,I422] [98,I4(1)22]
hP [143,P3] [144,P3(1)] [145,P3(2)] [149,P312] [150,P321] [151,P3(1)12]
[152,P3(1)21] [153,P3(2)12] [154,P3(2)21] [168,P6] [169,P6(1)]
[170,P6(5)] [171,P6(2)] [172,P6(4)] [173,P6(3)] [177,P622]
[178,P6(1)22] [179,P6(5)22] [180,P6(2)22] [181,P6(4)22] [182,P6(3)22]
hR [146,R3] [155,R32]
cP [195,P23] [198,P2(1)3] [207,P432] [208,P4(2)32] [212,P4(3)32]
[213,P4(1)32]
cF [196,F23] [209,F432] [210,F4(1)32]
cI [197,I23] [199,I2(1)3] [211,I432] [214,I4(1)32]
Subgroup and supergroup relations of these space groups
Compiled from International Tables for Crystallography (2006) Vol. A1 (Wiley). Simply put, for each space group, a maximum translationengleiche subgroup has lost a single type of symmetry, and a minimum translationengleiche supergroup has gained a single symmetry type. Example: P222 is a supergroup of P2, and a subgroup of P422 (and P4222 and P23). Of course the sub-/supergroup relation is recursive, which is why P1 is also a (sub-)subgroup of P222 (but not a maximum translationengleiche subgroup). The table below does not show other types of relations, e.g. non-isomorphic klassengleiche supergroups which may result e.g. from centring translations, because I find them less relevant in space group determination.
The table is relevant because in particular (perfect) twinning adds a symmetry type, and leads to an apparent space group which is the supergroup of the true space group.
| spacegroup number | maximum translationengleiche subgroup | minimum translationengleiche supergroup | spacegroup name |
|---|---|---|---|
| 1 | - | 3, 4, 5, 143, 144, 145, 146 | P 1 |
| 3 | 1 | 16, 17, 18, 21, 75, 77, 168, 171, 172 | P 2 |
| 4 | 1 | 17, 18, 19, 20, 76, 78, 169, 170, 173 | P 21 |
| 5 | 1 | 20, 21, 22, 23, 24, 79, 80, 149, 150, 151, 152, 153, 154, 155 | C2 |
| 16 | 3 | 89, 93, 195 | P 2 2 2 |
| 17 | 3, 4 | 91, 95 | P 2 2 21 |
| 18 | 3, 4 | 90, 94 | P 21 21 2 |
| 19 | 4 | 92, 96, 198 | P 21 21 21 |
| 20 | 4, 5 | 91, 92, 95, 96, 178, 179, 182 | C 2 2 21 |
| 21 | 3, 5 | 89, 90, 93, 94, 177, 180, 181 | C 2 2 2 |
| 22 | 5 | 97, 98, 196 | F 2 2 2 |
| 23 | 5 | 97, 197 | I 2 2 2 |
| 24 | 5 | 98, 199 | I 21 21 21 |
| 75 | 3 | 89, 90 | P 4 |
| 76 | 4 | 91, 92 | P 41 |
| 77 | 3 | 93, 94 | P 42 |
| 78 | 4 | 95, 96 | P 43 |
| 79 | 5 | 97 | I 4 |
| 80 | 5 | 98 | I 41 |
| 89 | 16, 21, 75 | 207 | P 4 2 2 |
| 90 | 18, 21, 75 | - | P 4 21 2 |
| 91 | 17, 20, 76 | - | P 41 2 2 |
| 92 | 19, 20, 76 | 213 | P 41 21 2 |
| 93 | 16, 21, 77 | 208 | P 42 2 2 |
| 94 | 18, 21, 77 | 93, 97 | P 42 21 2 |
| 95 | 17, 20, 78 | - | P 43 2 2 |
| 96 | 19, 20, 78 | 212 | P 43 21 2 |
| 97 | 22, 23, 79 | 209, 211 | I 4 2 2 |
| 98 | 22, 24, 80 | 210, 214 | I 41 2 2 |
| 143 | 1 | 149, 150, 168, 173 | P 3 |
| 144 | 1 | 151, 152, 169, 172 | P 31 |
| 145 | 1 | 153, 154, 170, 171 | P 32 |
| 146 | 1 | 155, 195, 196, 197, 198, 199 | R 3 |
| 149 | 5, 143 | 177, 182 | P 3 1 2 |
| 150 | 5, 143 | 177, 182 | P 3 2 1 |
| 151 | 5, 144 | 178, 181 | P 31 1 2 |
| 152 | 5, 144 | 178, 181 | P 31 2 1 |
| 153 | 5, 145 | 179, 180 | P 32 1 2 |
| 154 | 5, 145 | 179, 180 | P 32 2 1 |
| 155 | 5, 146 | 207, 208, 209, 210, 211, 212, 213, 214 | R 3 2 |
| 168 | 3, 143 | 177 | P 6 |
| 169 | 4, 144 | 178 | P 61 |
| 170 | 4, 145 | 179 | P 65 |
| 171 | 3, 145 | 180 | P 62 |
| 172 | 3, 144 | 181 | P 64 |
| 173 | 4, 143 | 182 | P 63 |
| 177 | 21, 149, 150, 168 | - | P 6 2 2 |
| 178 | 20, 151, 152, 169 | - | P 61 2 2 |
| 179 | 20, 153, 154, 170 | - | P 65 2 2 |
| 180 | 21, 153, 154, 171 | - | P 62 2 2 |
| 181 | 21, 151, 152, 172 | - | P 64 2 2 |
| 182 | 20, 149, 150, 173 | - | P 63 2 2 |
| 195 | 16, 146 | 207, 208 | P 2 3 |
| 196 | 22, 146 | 209, 210 | F 2 3 |
| 197 | 23, 146 | 211 | I 2 3 |
| 198 | 19, 146 | 212, 213 | P 21 3 |
| 199 | 24, 146 | 214 | I 21 3 |
| 207 | 89, 155, 195 | - | P 4 3 2 |
| 208 | 93, 155, 195 | - | P 42 3 2 |
| 209 | 97, 155, 196 | - | F 4 3 2 |
| 210 | 98, 155, 196 | - | F 41 3 2 |
| 211 | 97, 155, 197 | - | I 4 3 2 |
| 212 | 96, 155, 198 | - | P 43 3 2 |
| 213 | 92, 155, 198 | - | P 41 3 2 |
| 214 | 98, 155, 199 | - | I 41 3 2 |
(compare International Tables for Crystallography Vol A (2006), figure 10.1.3.2)
Space group selected by XDS: ambiguous with respect to enantiomorph and screw axes
In case of a crystal with an unknown space group (SPACE_GROUP_NUMBER=0 in XDS.INP), XDS (since version June 2008) helps the user in determination of the correct space group, by suggesting possible space groups compatible with the Laue symmetry and Bravais type of the data, and by calculating the Rmeas for these space groups.
XDS (or rather, the CORRECT step) makes an attempt to pick the correct space group automatically: it chooses the space group (or rather: Laue point group) which has the highest symmetry (thus yielding the lowest number of unique reflections) and still a tolerable Rmeas compared to the Rmeas the data have in any space group (which is most likely a low-symmetry space group - often P1).
In some cases the automatic choice is the correct one, and re-running the CORRECT step is then not necessary. However, neither the correct enantiomorph nor screw axes (see below) are determined automatically by XDS. Pointless is a very good program (usually better than CORRECT) to suggest possible space group (and alternatives). See also Notes, and checking the CORRECT assignment with pointless (below).
Space group selected by user
In case the space group selected by XDS should be incorrect, the resulting list (in CORRECT.LP) should give the user enough information to pick the correct space group herself (alternatives are in the big table above!). The user may then put suitable lines with SPACE_GROUP_NUMBER=, UNIT_CELL_CONSTANTS= into XDS.INP and re-run the CORRECT step to obtain the desired result. (The REIDX= line is no longer required; XDS figures the matrix out.)
Influencing the selection by XDS
keywords and parameters
The automatic choice is influenced by a number of decision constants that may be put into XDS.INP but which have defaults as indicated below:
- MAX_CELL_AXIS_ERROR= 0.03 ! relative deviation of unconstrained cell axes from those constrained by lattice symmetry
- MAX_CELL_ANGLE_ERROR= 3.0 ! degrees deviation of unconstrained cell angles from those constrained by lattice symmetry
- TEST_RESOLUTION_RANGE= 10.0 5.0 ! resolution range for calculation of Rmeas
- MIN_RFL_Rmeas= 50 ! at least this number of reflections are required
- MAX_FAC_Rmeas= 2.0 ! factor to multiply the lowest Rmeas with to still be acceptable
The user may experiment with adjusting these values to make the automatic mode of space group determination more successful. For example, if the crystal diffracts weakly, all Rmeas values will be high and no valid decision can be made. In this case, I suggest to use e.g.
TEST_RESOLUTION_RANGE= 50.0 10.0
dealing with alternative indexing
There are two ways to have XDS choose an indexing consistent with some other dataset:
- using REFERENCE_DATA_SET= ! see also REFERENCE_DATA_SET
- using UNIT_CELL_A-AXIS=, UNIT_CELL_B-AXIS=, UNIT_CELL_C-AXIS= from a previous data collection run with the same crystal
One can also manually force a specific indexing, using the REIDX= keyword, but this is error-prone.
See also http://www.ccp4.ac.uk/html/reindexing.html
Screw axes
The current version makes no attempt to find out about screw axes. It is assumed that the user checks the table in CORRECT.LP entitled REFLECTIONS OF TYPE H,0,0 0,K,0 0,0,L OR EXPECTED TO BE ABSENT (*), and identifies whether the intensities follow the rules
- a two-fold screw axis along an axis in reciprocal space (theoretically) results in zero intensity for the odd-numbered (e.g. 0,K,0 with K = 2*n + 1) reflections, leaving the reflections of type 2*n as candidates for medium to strong reflections (they don't have to be strong, but they may be strong!).
- similarly, a three-fold screw axis (theoretically) results in zero intensity for the reflections of type 3*n+1 and 3*n+2, and allows possibly strong 3*n reflections . 31 and 32 cannot be distinguished - they are enantiomorphs.
- analogously for four-fold screw axes: for H=0, K=0 reflections, 41 and 43 screws yield the rule L=4*n, and 42 yields the rule L=2*n .
- analogously for six-fold screw axes: for H=0, K=0 reflections, 61 and 65 screws yield the rule L=6*n, 62 and 64 yield the rule L=3*n, and 63 yields the rule L=2*n .
Once screw axes have been deduced from the patterns of intensities along H,0,0 0,K,0 0,0,L , the resulting space group should be identified in the lists of space group numbers printed in IDXREF.LP and CORRECT.LP, and the CORRECT step can be re-run. Those reflections that should theoretically have zero intensity are then marked with a "*" in the table. In practice, they should be (hopefully quite) weak, or even negative.
Notes
- There is still the old Wiki page Old way of Space group determination
- To prevent XDS from trying to find a better space group than the one with the lowest Rmeas (often P1), you could just use MAX_FAC_Rmeas= 1.0
- Space group determination is not a trivial task. There is a number of difficulties that arise -
- ambiguity of hand of screw axis (e.g. 31 versus 32, or 62 versus 64) - see the table above!
- twinning may make a low-symmetry space group look like a high-symmetry one
- pointless helps with identification of screw axes, and its identification of Laue group is more sensitive than that implemented in CORRECT. However it is not fail-safe, and cannot tell the correct enantiomorph.
- only when the structure is satisfactorily refined can the chosen space group be considered established and correct. Until then, it is just a hypothesis and its alternatives (see the table) should be kept in mind.
An example
The following example shows the difficulty in deciding between higher and lower symmetry.
IDXREF reports:
LATTICE- BRAVAIS- QUALITY UNIT CELL CONSTANTS (ANGSTROEM & DEGREES)
CHARACTER LATTICE OF FIT a b c alpha beta gamma
* 44 aP 0.0 74.0 78.6 124.0 108.2 105.1 90.4
* 31 aP 3.5 74.0 78.6 124.0 71.8 74.9 90.4
* 41 mC 7.4 235.7 78.6 74.0 90.4 106.1 89.7
* 37 mC 34.8 239.6 74.0 78.6 90.4 109.0 87.8
* 42 oI 38.7 74.0 78.6 226.6 90.2 92.2 90.4
14 mC 122.5 107.6 108.3 124.0 92.8 114.1 86.6
...
so oI (space groups 23=I222 or 24=I212121) or the two mC (space group 5=C2) are possibilities for indexing, and of course P1. Integration in P1 (because of SPACE_GROUP_NUMBER=0 in XDS.INP) results in the following table for space group determination in CORRECT:
SPACE-GROUP UNIT CELL CONSTANTS UNIQUE Rmeas COMPARED LATTICE-
NUMBER a b c alpha beta gamma CHARACTER
1 74.1 78.6 124.0 71.8 74.8 90.4 6959 22.5 5309 31 aP
* 23 74.1 78.6 226.6 90.0 90.0 90.0 2735 39.5 9533 42 oI
5 235.7 78.6 74.1 90.0 106.1 90.0 3922 31.0 8346 41 mC
5 239.6 74.1 78.6 90.0 109.0 90.0 5111 30.7 7157 37 mC
1 74.1 78.6 124.0 108.2 105.2 90.4 6959 22.5 5309 44 aP
The automatic choice gave 23 as a result. This is because the associated Rmeas (39.5) is lower than MAX_FAC_RMEAS * (lowest Rmeas) = 2 * 22.5 = 45.0. However, the C2 values of 30.7 and 31.0 are significantly lower still, and the question needs further investigation.
checking the CORRECT assignment with pointless
pointless XDS_ASCII.HKL gives:
Analysing rotational symmetry in lattice group I 4/m m m
----------------------------------------------
<!--SUMMARY_BEGIN-->
Scores for each symmetry element
Nelmt Lklhd Z-cc CC N Rmeas Symmetry & operator (in Lattice Cell)
1 0.856 7.77 0.78 9368 0.224 identity
2 0.139 2.98 0.30 6376 0.894 2-fold l ( 0 0 1) {-h,-k,l}
3 0.842 8.02 0.80 18679 0.234 ** 2-fold k ( 0 1 0) {-h,k,-l}
4 0.148 3.10 0.31 5858 1.254 2-fold h ( 1 0 0) {h,-k,-l}
5 0.073 1.25 0.13 12111 1.077 2-fold ( 1-1 0) {-k,-h,-l}
6 0.071 1.11 0.11 12293 1.152 2-fold ( 1 1 0) {k,h,-l}
7 0.068 0.83 0.08 24758 1.084 4-fold l ( 0 0 1) {-k,h,l}{k,-h,l}
and this shows that actually there is only one instead of three two-fold axes, despite the fact that CORRECT auto-chose the orthorhombic system and possibly even rejected some reflections that do not fit it well. Pointless then re-indexes to I2 and suggests:
Best Solution: space group I 1 2 1
Reindex operator: [h,k,l]
Laue group probability: 0.763
Systematic absence probability: 1.000
Total probability: 0.763
Space group confidence: 0.687
Laue group confidence 0.687
Unit cell: 78.05 80.78 235.63 90.00 90.00 90.00
50.67 to 3.73 - Resolution range used for Laue group search
50.67 to 3.08 - Resolution range in file, used for systematic absence check
Number of batches in file: 120
The data do not appear to be twinned, from the L-test
$$ <!--SUMMARY_END-->
HKLIN spacegroup: I 2 2 2 body-centred orthorhombic
$TEXT:Warning:$$ $$
The input crystal system is body-centred orthorhombic
(Cell: 78.05 80.78 235.63 90.00 90.00 90.00)
The crystal system chosen for output is body-centred monoclinic
(Cell: 78.05 80.78 235.63 90.00 90.00 90.00)
$TEXT:Warning:$$ $$
WARNING:
WARNING:
The chosen output crystal system is different from that used for integration of the input file(s).
You should rerun the integration in the chosen crystal system because the cell constraints differ
$$
Filename:
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
Final point group choice has alternative indexing possibilities
Alternative indexing possibilities are marked '*' if the cells are
too different at the maximum resolution
CellDifference(A) ReindexOperator
1 0.0 [h,k,l]
2 0.0 [h,-k,-l]
3 8.0 * [k,-h,l]
4 8.0 * [-k,h,l]
At the bottom of the output, we see that other indexing possibilities exist. That means that one has to be very careful when merging crystals from this project, and that there may also be the possibility of twinning.
In this particular case, CORRECT chose the wrong symmetry, whereas pointless identified the correct symmetry elements. I have also seen cases where pointless mis-identified the symmetry, usually on the side of too high symmetry. To avoid mistakes in space group identification, it is absolutely crucial to read and understand the tables that the programs print.
a more sensible way to run pointless
pointless should really be run with the "SETTING SYMMETRY-BASED" option. When doing that, the output changes to
Best Solution: space group C 1 2 1 Reindex operator: [h+l,k,-h] Laue group probability: 0.807 Systematic absence probability: 1.000 Total probability: 0.807 Space group confidence: 0.734 Laue group confidence 0.734 Unit cell: 248.53 80.86 78.08 90.00 108.31 90.00 40.76 to 3.80 - Resolution range used for Laue group search 40.76 to 3.08 - Resolution range in file, used for systematic absence check Number of batches in file: 120 The data do not appear to be twinned, from the L-test $$ <!--SUMMARY_END--> HKLIN spacegroup: I 2 2 2 body-centred orthorhombic $TEXT:Warning:$$ $$ The input crystal system is body-centred orthorhombic (Cell: 78.08 80.86 235.95 90.00 90.00 90.00) The crystal system chosen for output is C-centred monoclinic (Cell: 248.53 80.86 78.08 90.00 108.31 90.00) $TEXT:Warning:$$ $$ WARNING: WARNING: The chosen output crystal system is different from that used for integration of the input file(s). You should rerun the integration in the chosen crystal system because the cell constraints differ $$
so the spacegroup given is the more normal C2 setting (instead of A2 or I2). Unfortunately, pointless does not seem to print out the table of "alternative indexing possibilities" in this mode - but possibly the table is only printed in the I2 case because the beta angle comes out as 90.0°.
not biasing the cell parameters, and avoiding premature outlier rejection
CORRECT can easily be forced not to assign a spacegroup, and consequently will not reject outliers based on a too high symmetry assignment. To this end one simply supplies space group P1 and its unit cell:
SPACE_GROUP_NUMBER=1 UNIT_CELL_CONSTANTS= 74.1 78.6 124.0 108.2 105.2 90.4
in XDS.INP and re-runs CORRECT. That gives an unbiased XDS_ASCII.HKL, with no angles set to 90°. pointless with SETTING SYMMETRY_BASED then gives
Best Solution: space group C 1 2 1 Reindex operator: [-k-2l,-k,-h] Laue group probability: 0.849 Systematic absence probability: 1.000 Total probability: 0.849 Space group confidence: 0.770 Laue group confidence 0.770 Unit cell: 233.70 78.42 73.22 90.31 105.34 89.77
and without SETTING SYMMETRY_BASED
Best Solution: space group I 1 2 1 Reindex operator: [-h,-k,h+k+2l] Laue group probability: 0.838 Systematic absence probability: 1.000 Total probability: 0.838 Space group confidence: 0.753 Laue group confidence 0.753 Unit cell: 233.70 78.42 73.22 90.31 105.34 89.77
the dreadful I2 space group nomenclature (and I have no idea why the probability and confidence values are worse than in C2). But anyway this shows that there are two non-equivalent ways to index the same lattice!
final steps
Finally, XDS (but only JOB=DEFPIX INTEGRATE CORRECT) should be re-run with
SPACE_GROUP_NUMBER=5 UNIT_CELL_CONSTANTS= 233.70 78.42 73.22 90 105.34 90
because this enforces just the correct cell constraints.