1RQW: Difference between revisions
mNo edit summary |
|||
| Line 5: | Line 5: | ||
Prior to flash cooling in liquid nitrogen at 100 K the crystals were soaked for a few seconds in a solution containing 1 M sodium bromide and 25% (v/v) glycerol. | Prior to flash cooling in liquid nitrogen at 100 K the crystals were soaked for a few seconds in a solution containing 1 M sodium bromide and 25% (v/v) glycerol. | ||
The peak and inflection datasets (360 images each) are available from the [http://www.embl-hamburg.de/Xray_Tutorial/ EMBL Hamburg] website (alternatively from [http://www.mx.bessy.de/bessy-ws/experiment2.html]). The XDS data reductions and (some of the) phasing calculations are [ftp:// | The peak and inflection datasets (360 images each) are available from the [http://www.embl-hamburg.de/Xray_Tutorial/ EMBL Hamburg] website (alternatively from [http://www.mx.bessy.de/bessy-ws/experiment2.html]). The XDS data reductions and (some of the) phasing calculations are [ftp://strucbio.biologie.uni-konstanz.de//pub/xds-datared/1rqw here]. | ||
Data reduction was performed starting with the [[XDS.INP#MarCDD_225mm_.40_SLS.2C_BL_X06SA|template for the MarCCD 225]] detector, changing | Data reduction was performed starting with the [[XDS.INP#MarCDD_225mm_.40_SLS.2C_BL_X06SA|template for the MarCCD 225]] detector, changing | ||
Revision as of 17:06, 19 December 2019
Thaumatin Br soak: structure solution from either 2-wavelength MAD, or from single wavelength SAD
Thaumatin is a sweet-tasting protein of 207 residues that crystallizes in P41212 with a monomer in the asymmetric unit.
Prior to flash cooling in liquid nitrogen at 100 K the crystals were soaked for a few seconds in a solution containing 1 M sodium bromide and 25% (v/v) glycerol.
The peak and inflection datasets (360 images each) are available from the EMBL Hamburg website (alternatively from [1]). The XDS data reductions and (some of the) phasing calculations are here.
Data reduction was performed starting with the template for the MarCCD 225 detector, changing
ORGX=1533 ORGY=1536 DETECTOR_DISTANCE=200 OSCILLATION_RANGE=0.5 X-RAY_WAVELENGTH=0.91878 ! peak ! X-RAY_WAVELENGTH=0.9196 ! inflection NAME_TEMPLATE_OF_DATA_FRAMES=../peak/thau_peak_1_???.mccd DIRECT TIFF ! NAME_TEMPLATE_OF_DATA_FRAMES=../inf/thau_inf_1_???.mccd DIRECT TIFF DATA_RANGE=1 360 SPACE_GROUP_NUMBER=92 UNIT_CELL_CONSTANTS= 57.8 57.8 150. 90 90 90 FRIEDEL'S_LAW=FALSE VALUE_RANGE_FOR_TRUSTED_DETECTOR_PIXELS=9000. 30000.
SAD: Peak data alone
G. Sheldrick recommends: "For iodide soaks, a good rule of thumb is to start with a number of iodide sites equal to the number of amino-acids in the asymmetric unit divided by 15." Bromine should behave similarly, so we expect roughly 14 sites.
manual structure solution using hkl2map and buccaneer
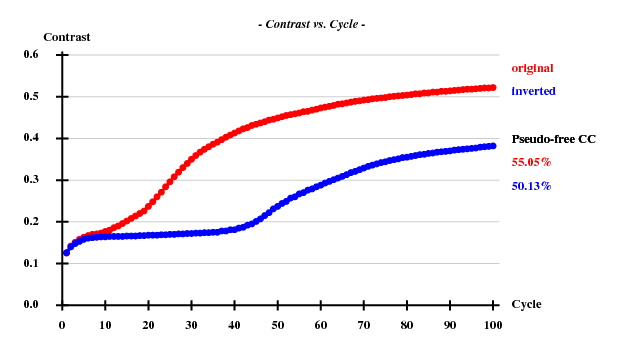
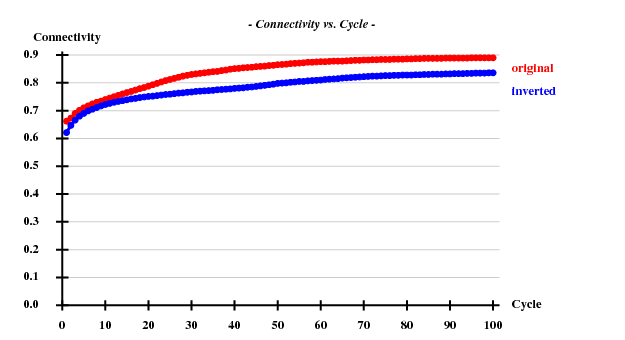
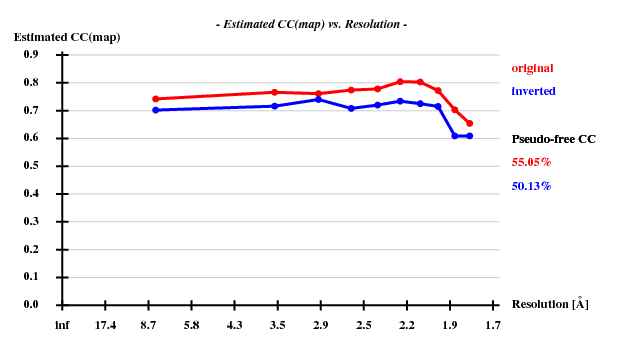
The structure was "solved" using the hkl2map GUI. Based on the statistics
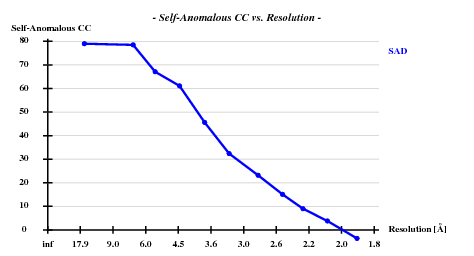
Resl. Inf - 8.0 - 6.0 - 5.0 - 4.0 - 3.5 - 3.0 - 2.6 - 2.4 - 2.2 - 2.0 - 1.80 N(data) 346 425 519 1144 1119 1964 2834 2163 3043 4326 6351 Chi-sq 0.71 1.08 0.44 1.03 1.17 1.06 1.06 1.09 1.10 1.08 1.10 I/sig 47.4 41.8 45.5 50.7 46.1 36.0 22.6 15.4 12.0 8.0 4.1 %Complete 94.8 98.6 99.0 99.5 99.6 99.6 99.8 99.8 99.8 99.8 99.6 <d"/sig> 3.13 2.58 2.13 1.90 1.52 1.36 1.16 1.04 0.96 0.91 0.85 CC(anom) 79.0 78.5 67.1 61.1 45.6 32.4 23.2 15.1 9.0 3.8 -3.6
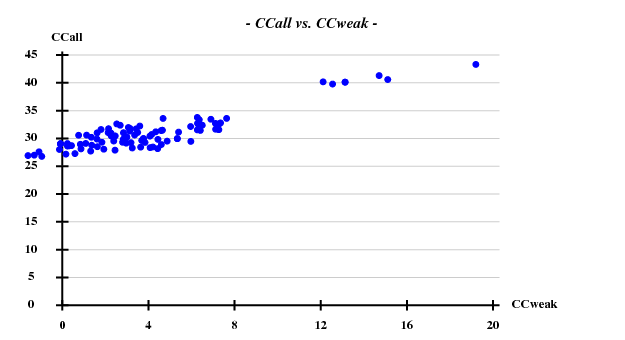
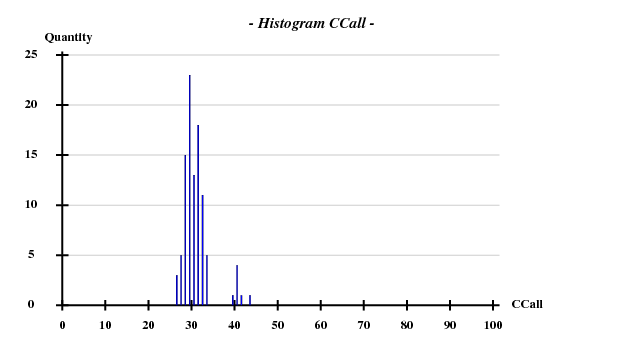
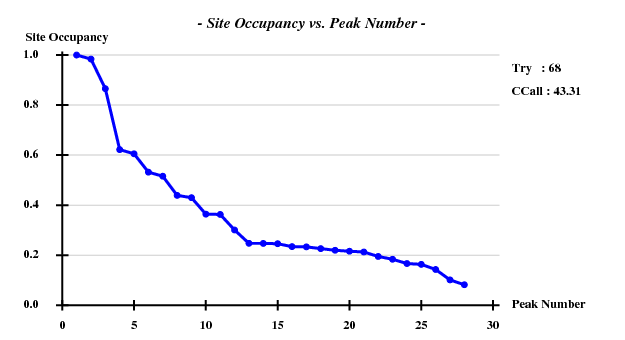
I decided to use 3.3 Å as a suitable cutoff for solving the substructure, and to let SHELXD search for 20 Br atoms. SHELXD then found a convincing solution:
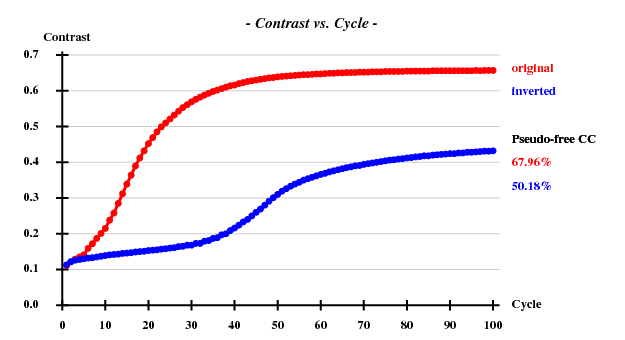
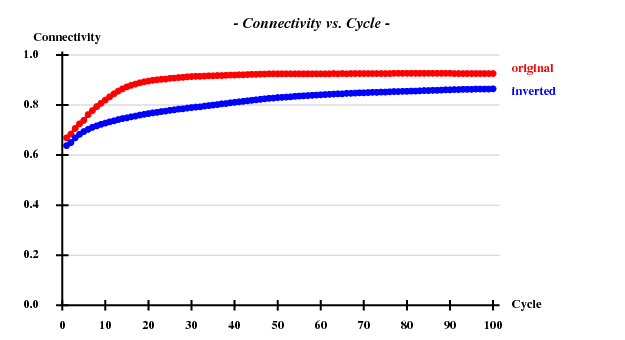
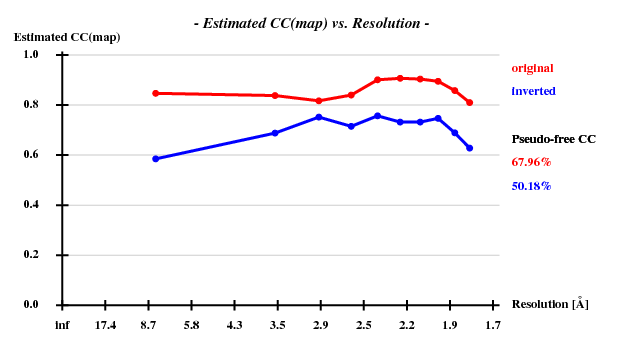
and SHELXE produced useful phases:
Using ccp4i, I imported the .phs file from SHELXE, gave it to ccp4i's buccaneer- autobuild/refine task using default parameters, and obtained in the third cycle from buccaneer 0.9.9:
202 residues were built in 9 chains, the longest having 57 residues. 138 residues were sequenced, after pruning.
Refmac5 refined this to R/R_free of 39.0%/42.4% which shows that the structure is essentially solved.
automatic structure solution using Auto-Rickshaw (http://www.embl-hamburg.de/Auto-Rickshaw/)
This uses Santosh Panjikar's script DPS2AR.csh version 1.04:
DPS2AR.csh datafile1 thau-peak-I.mtz keeps world ver completeversion nres 207 nsm 20 masu 1 meth SAD SG P41212 \
email kay.diederichs@uni-konstanz.de sequencefile 1rqw.seq
thau-peak-I.mtz was obtained in two steps:
a) generating temp.hkl from XDS_ASCII.HKL with the following XDSCONV.INP:
INPUT_FILE=XDS_ASCII.HKL OUTPUT_FILE=temp.hkl CCP4_I ! Warning: do _not_ name this file "temp.mtz" ! FRIEDEL'S_LAW=FALSE ! default is FRIEDEL'S_LAW=TRUE
b) running the conversion to thau-peak-I.mtz as indicated by the XDSCONV screen output.
1rqw.seq is
ATFEIVNRCS YTVWAAASKG DAALDAGGRQ LNSGESWTIN VEPGTKGGKI WARTDCYFDD SGSGICKTGD CGGLLRCKRF GRPPTTLAEF SLNQYGKDYI DISNIKGFNV PMDFSPTTRG CRGVRCAADI VGQCPAKLKA PGGGCNDACT VFQTSEYCCT TGKCGPTEYS RFFKRLCPDA FSYVLDKPTT VTCPGSSNYR VTFCPTA
The result of this (see the Auto-Rickshaw logfile) is a model that only lacks residues 1, 82, 83, 207, and which has a "core RMS" (from coot's SSM superpose) of 0.14 Å against 1rqw (which is a 1.05 Å structure).
It couldn't be simpler than that. Thanks, Santosh!
SAD: Inflection data alone
This is of course much more difficult, because the anomalous data are weaker (and as this is the second dataset collected from the crystal maybe there is already a little bit of radiation damage).
A lot of care was put into the XDS data reduction - many of the ideas that can be found in this wiki were employed to get as good data as possible.
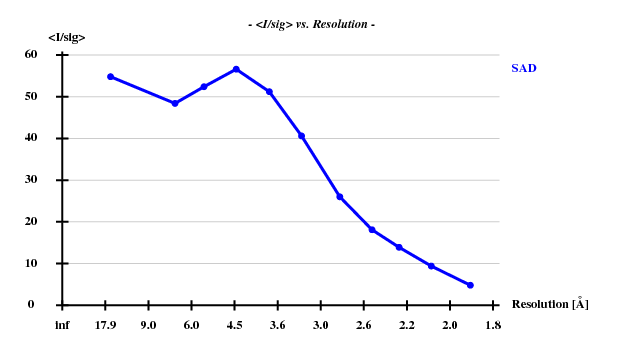
The result is that the anomalous signal is useful - this is evident from the SHELXC statistics:
Resl. Inf - 8.0 - 6.0 - 5.0 - 4.0 - 3.5 - 3.0 - 2.6 - 2.4 - 2.2 - 2.0 - 1.80 N(data) 346 427 519 1142 1123 1972 2830 2173 3043 4340 6339 I/sig 54.8 48.4 52.4 56.6 51.2 40.6 26.0 18.1 13.9 9.4 4.8 %Complete 94.8 98.6 99.0 99.5 99.6 99.6 99.8 99.8 99.8 99.8 99.8 <d"/sig> 2.59 2.18 1.86 1.58 1.29 1.18 1.04 0.95 0.91 0.88 0.81
This time I tried to solve the substructure at 3.6 Å resolution, again 20 sites -
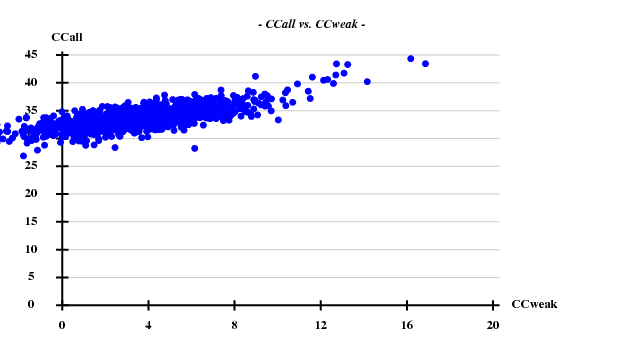
 So there are indeed much less good trials:
So there are indeed much less good trials:
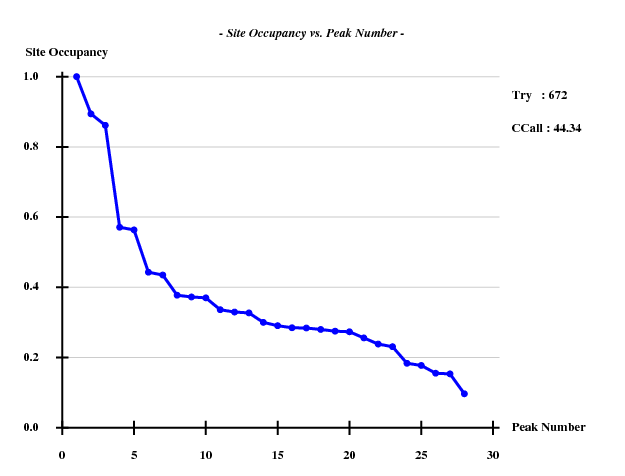
SHELXE was run with 53.5% solvent:
The structure can be solved from here, with Arp/Warp and probably also with buccaneer/refmac.
automatic structure solution using Auto-Rickshaw (http://www.embl-hamburg.de/Auto-Rickshaw/)
In the same way as with the peak data, Auto-Rickshaw was started. Please see the Auto-Rickshaw logfile.
The structure could be solved but Arp/Warp had a difficult start - take a look at the Arp/Warp logfile! The final model lacks residues 1, 85, 205, 206, 207 and the "core RMS" of SSM superpose, against 1rqw, is 0.15 Å (better than I would expect).
It should also be noted that the structure can not be solved when searching for 15 or 28 sites - rather, 20 appears to be the magic number.
Once again, great job, Santosh (and of course those people who wrote the programs that Auto-Rickshaw uses)!