Spot2pdb: Difference between revisions
No edit summary |
No edit summary |
||
| Line 18: | Line 18: | ||
and produces a CCP4 map file with pixel contents mapped to reciprocal space, to 6Å. | and produces a CCP4 map file with pixel contents mapped to reciprocal space, to 6Å. | ||
Notes | |||
== Examples == | |||
[[File:insu6A.png]] | |||
Indexed spot positions are yellow, un-indexed ones red. | |||
[[File:insu6AplusED.png]] | |||
Same, but with pseudo electron density. | |||
== Notes == | |||
* the mapping used by <code>dials.rs_mapper</code> uses the (unrefined) frame header values, and possibly assumptions (as implemented in <code>iotbx.detectors</code>) about geometry aspects, like ROTATION_AXIS, not stored in the header. On the other hand, XPARM.XDS may have different assumptions about geometry aspects, and uses the geometry as refined by IDXREF. Ideally, these two mappings should agree. | * the mapping used by <code>dials.rs_mapper</code> uses the (unrefined) frame header values, and possibly assumptions (as implemented in <code>iotbx.detectors</code>) about geometry aspects, like ROTATION_AXIS, not stored in the header. On the other hand, XPARM.XDS may have different assumptions about geometry aspects, and uses the geometry as refined by IDXREF. Ideally, these two mappings should agree. | ||
* Since <code>dials.rs_mapper</code> uses the STARTING_ANGLE information from the frame headers, the mappings can only agree if <code>XDS.INP</code> also has this item. [[generate_XDS.INP]] versions since 0.82 write <code>XDS.INP</code> with STARTING_ANGLE information; earlier versions don't. | * Since <code>dials.rs_mapper</code> uses the STARTING_ANGLE information from the frame headers, the mappings can only agree if <code>XDS.INP</code> also has this item. [[generate_XDS.INP]] versions since 0.82 write <code>XDS.INP</code> with STARTING_ANGLE information; earlier versions don't. | ||
* if no <code>XPARM.XDS</code> exists, one may use [ftp://turn5.biologie.uni-konstanz.de/pub/linux_bin/generate_XPARM.XDS XDS.INP as a source of geometry information]. | * if no <code>XPARM.XDS</code> exists, one may use [ftp://turn5.biologie.uni-konstanz.de/pub/linux_bin/generate_XPARM.XDS XDS.INP as a source of geometry information] for <code>spot2pdb</code>. | ||
* | * <code>coot</coot> connects atoms that are close, have residue numbers differing by at most 1, and are in the same chain, with bonds (lines). The program makes an attempt to avoid such lines by cycling through chain names; the proper solution would be <code>coot</code>'s ''display objects''. | ||
Revision as of 18:17, 6 March 2018
spot2pdb,[1] is a jiffy that serves to create pseudo-PDB files for visualization of reciprocal space, based on SPOT.XDS and XPARM.XDS. The "atom" positions in the pseudo-PDB files are actually reflections positions.
The usage is simple: just run
spot2pdb
in a XDS directory containing SPOT.XDS and XPARM.XDS. The program then creates SPOT-indexed.pdb and SPOT-notindexed.pdb, depending on whether SPOT.XDS has indices attached to spot positions or not (or just 0 0 0). Thus it is useful directly after COLSPOT, but even more after IDXREF. Reflections of SPOT-indexed.pdb can be clicked, and have (atomname residuename sequencenumber) corresponding to (H K L).
By default, the program creates pseudo-PDB files with reflections up to 6Å. The maximum resolution can be adjusted with the -r option.
Visualization can then be achieved with
coot SPOT-*.pdb
Since there exist duplicate residue numbers in SPOT-indexed.pdb, one needs a ~/.coot with the line
(allow-duplicate-sequence-numbers)
Even better visualization of the raw data and their abstraction as reflection positions is achieved with
coot --pdb SPOT-*.pdb --map rs_mapper_output.ccp4
provided that dials.rs_mapper is run beforehand. The latter is as easy as
dials.rs_mapper /path/to/reflection/files.ext
and produces a CCP4 map file with pixel contents mapped to reciprocal space, to 6Å.
Examples
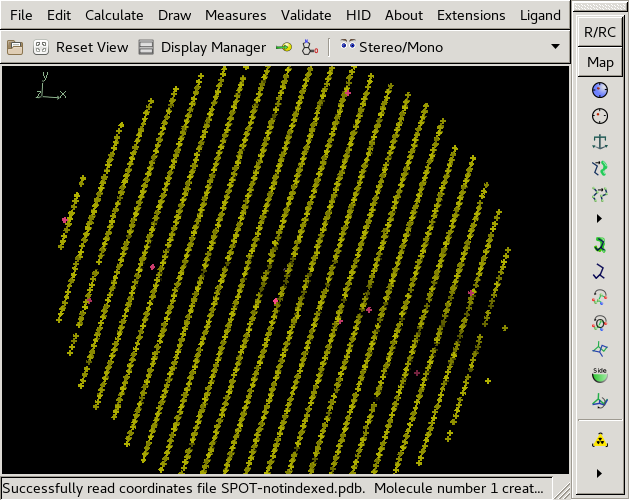 Indexed spot positions are yellow, un-indexed ones red.
Indexed spot positions are yellow, un-indexed ones red.
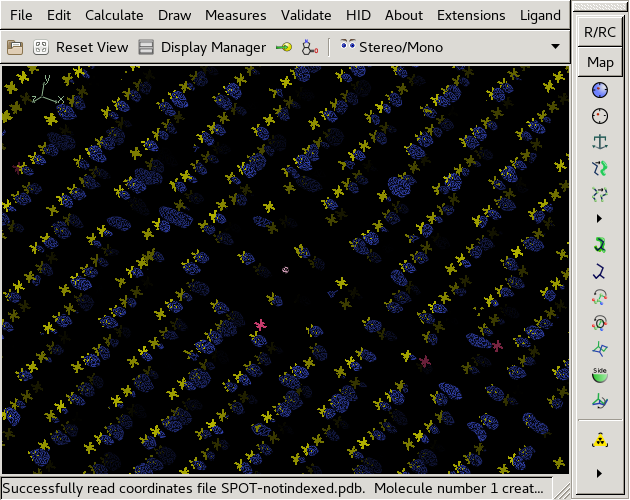 Same, but with pseudo electron density.
Same, but with pseudo electron density.
Notes
- the mapping used by
dials.rs_mapperuses the (unrefined) frame header values, and possibly assumptions (as implemented iniotbx.detectors) about geometry aspects, like ROTATION_AXIS, not stored in the header. On the other hand, XPARM.XDS may have different assumptions about geometry aspects, and uses the geometry as refined by IDXREF. Ideally, these two mappings should agree. - Since
dials.rs_mapperuses the STARTING_ANGLE information from the frame headers, the mappings can only agree ifXDS.INPalso has this item. generate_XDS.INP versions since 0.82 writeXDS.INPwith STARTING_ANGLE information; earlier versions don't. - if no
XPARM.XDSexists, one may use XDS.INP as a source of geometry information forspot2pdb. coot</coot> connects atoms that are close, have residue numbers differing by at most 1, and are in the same chain, with bonds (lines). The program makes an attempt to avoid such lines by cycling through chain names; the proper solution would becoot's display objects.