SSX: Difference between revisions
more PNGs |
No edit summary |
||
| Line 159: | Line 159: | ||
[[File:1g1c-94.png]] | [[File:1g1c-94.png]] | ||
(If the space group were correct, the result of xscale_isocluster should look similar to this: | |||
[[File:Lyso-xscale-isocluster.png]] | |||
which is from a lysozyme SSX data collection performed at the SLS; outliers are labelled. In this case, the data are truely tetragonal.) | |||
We must now investigate whether the data have lower than tetragonal symmetry. | We must now investigate whether the data have lower than tetragonal symmetry. | ||
| Line 172: | Line 178: | ||
cluster: 2 center: 6 elements: 49 core: 49 halo: 0 | cluster: 2 center: 6 elements: 49 core: 49 halo: 0 | ||
</pre> | </pre> | ||
and prepares XSCALE.1.INP (and XSCALE.2.INP) for further use. | and prepares XSCALE.1.INP (and XSCALE.2.INP) for further use (these two files collect the differently, but internally-consistently indexed XDS_ASCII.HKL files). | ||
coot iso.pdb | coot iso.pdb | ||
shows | shows | ||
| Line 178: | Line 184: | ||
[[File:Coot.png]] | [[File:Coot.png]] | ||
and thus reveals two well separated clouds | and thus reveals two well separated clouds, corresponding to the two possible indexing modes of the data in space group 19. | ||
Using XSCALE.1.INP with its 51 XDS_ASCII.HKL, and FRIEDEL'S_LAW=TRUE, we get | Using XSCALE.1.INP with its 51 XDS_ASCII.HKL, and FRIEDEL'S_LAW=TRUE, we get | ||
| Line 269: | Line 269: | ||
== Round 2: using the REFERENCE_DATA_SET == | == Round 2: using the REFERENCE_DATA_SET == | ||
The processing script integrate.rc is changed a bit, to a) use the REFERENCE_DATA_SET, b) prevent adjustment of variances by CORRECT (this should be done by XSCALE) , c) allow some radiation damage | The processing script integrate.rc is changed a bit, to a) use the REFERENCE_DATA_SET, b) prevent adjustment of variances by CORRECT (this should be done by XSCALE) , c) allow some radiation damage correction in XSCALE: | ||
<pre> | <pre> | ||
#!/bin/bash -f | #!/bin/bash -f | ||
Revision as of 11:53, 29 December 2016
This article deals with how to process serial synchrotron crystallography (SSX) data.
The particular data we are processing are artificial and were prepared by James Holton. The files Illuin_microfocus_minimal_00[1-3].tar.bz2 can be downloaded and the data and problem are described on his microfocus challenge page.
The challenges are
- partial data sets: each of the 100 data sets has only 3 frames of 1° oscillation
- strong radiation damage: the crystals decay to about 1/2 within these 3 frames
- the b and c axes are the same length, but the crystals are orthorhombic. This makes it difficult to index them consistently - it is wrong to just merge them, because that yields a pseudo-tetragonal merged data set.
Round 1: processing the data, and determining the space group
In order to be able to merge the data in XSCALE, we must ensure that they are all processed in the same space group, with similar cell parameters. Some exploratory processing (not shown) and averaging of cell parameters reveals that IDXREF finds a primitive lattice with one axis of 38.3 Å, and two with 79.1 Å; angles are 90°. The data go to 1.8 Å; beyond that, the intensities suddenly drop to 0 - presumably because James Holton simulated them only that far. Using the following as the processing script integrate.rc:
#!/bin/bash -f
for f in `seq 1 100`;
do
export OUT=wedge0`printf "%03d" $f`
export NAMES="$PWD/Illuin/microfocus/xtal"`printf "%03d" $f`"_1_00\?.img"
rm -rf $OUT
mkdir $OUT
cd $OUT
generate_XDS.INP $NAMES
sed -i s"/SPOT_RANGE=1 1/SPOT_RANGE=1 3/" XDS.INP
sed -i s"/SPACE_GROUP_NUMBER=0/SPACE_GROUP_NUMBER=1/" XDS.INP
sed -i s"/UNIT_CELL_CONSTANTS= 70 80 90/UNIT_CELL_CONSTANTS=38.3 79.1 79.1/" XDS.INP
sed -i s"/TRUSTED_REGION=0.0 1.2/TRUSTED_REGION=0 1/" XDS.INP
sed -i s"/INCLUDE_RESOLUTION_RANGE=50 0/INCLUDE_RESOLUTION_RANGE=99 1.8/" XDS.INP
/usr/local/bin/xds_par
cd ..
done
mkdir xscale
cd xscale
cat >XSCALE.INP <<eof
SPACE_GROUP_NUMBER= 1
UNIT_CELL_CONSTANTS= 38.3 79.1 79.1 90 90 90
OUTPUT_FILE=temp.ahkl
SAVE_CORRECTION_IMAGES=FALSE
FRIEDEL'S_LAW=TRUE
eof
find $PWD/../wedge* -name XDS_ASCII.HKL | awk '{print "INPUT_FILE=",$0;print "NBATCH=1 CORRECTIONS=ALL"}' >> XSCALE.INP
we obtain in P1
SUBSET OF INTENSITY DATA WITH SIGNAL/NOISE >= -3.0 AS FUNCTION OF RESOLUTION
RESOLUTION NUMBER OF REFLECTIONS COMPLETENESS R-FACTOR R-FACTOR COMPARED I/SIGMA R-meas CC(1/2) Anomal SigAno Nano
LIMIT OBSERVED UNIQUE POSSIBLE OF DATA observed expected Corr
8.03 3014 908 958 94.8% 44.5% 42.0% 2896 2.55 52.1% 65.0* 3 0.983 231
5.68 5502 1679 1788 93.9% 46.8% 42.5% 5239 2.50 54.8% 50.3* 6 1.001 390
4.64 6996 2164 2292 94.4% 47.5% 42.3% 6656 2.48 55.9% 68.4* 5 1.080 495
4.01 8079 2580 2735 94.3% 48.7% 42.5% 7591 2.38 57.3% 50.0* 2 1.106 557
3.59 9167 2904 3099 93.7% 52.1% 42.7% 8694 2.36 61.7% 43.6* -6 1.017 599
3.28 10276 3226 3397 95.0% 53.3% 43.3% 9728 2.35 62.8% 36.0* 1 1.104 708
3.03 11040 3472 3687 94.2% 54.5% 44.3% 10500 2.17 64.2% 44.4* 2 1.044 728
2.84 12022 3771 3977 94.8% 55.9% 47.2% 11424 1.97 65.8% 36.2* 3 0.999 835
2.68 12705 3985 4227 94.3% 58.5% 51.0% 12065 1.78 68.8% 37.8* -3 0.934 898
2.54 13370 4252 4489 94.7% 59.5% 56.2% 12670 1.61 70.5% 30.1* 4 0.887 869
2.42 14299 4505 4744 95.0% 62.4% 63.6% 13594 1.46 73.7% 30.2* -2 0.824 979
2.32 14835 4647 4915 94.5% 63.8% 70.0% 14083 1.35 75.1% 29.9* -2 0.765 1041
2.23 15599 4917 5181 94.9% 65.7% 72.6% 14809 1.31 77.5% 27.6* -1 0.756 1075
2.15 15888 4965 5272 94.2% 65.1% 78.6% 15117 1.28 76.9% 26.8* -2 0.708 1115
2.07 16872 5324 5601 95.1% 69.1% 88.1% 16035 1.14 81.6% 22.2* 3 0.687 1119
2.01 16856 5349 5649 94.7% 73.4% 92.5% 15988 1.06 86.5% 19.7* -3 0.673 1144
1.95 17842 5666 5976 94.8% 76.7% 105.9% 16959 0.97 90.8% 20.7* -8 0.606 1189
1.89 18102 5767 6069 95.0% 84.4% 127.9% 17152 0.85 99.9% 15.1* -1 0.590 1183
1.84 18633 5933 6256 94.8% 92.8% 162.0% 17667 0.72 109.8% 17.6* 0 0.533 1236
1.80 15519 5405 6479 83.4% 103.0% 194.1% 14280 0.58 122.7% 18.2* 1 0.503 940
total 256616 81419 86791 93.8% 54.3% 51.3% 243147 1.43 64.0% 64.6* 0 0.788 17331
and feed this to pointless:
pointless xdsin temp.ahkl
which tells us
Scores for each symmetry element
Nelmt Lklhd Z-cc CC N Rmeas Symmetry & operator (in Lattice Cell)
1 0.854 5.41 0.54 801 0.706 identity
2 0.842 4.62 0.46 785 0.819 ** 2-fold l ( 0 0 1) {-h,-k,l}
3 0.867 5.13 0.51 746 0.912 ** 2-fold k ( 0 1 0) {-h,k,-l}
4 0.837 5.64 0.56 735 0.807 ** 2-fold h ( 1 0 0) {h,-k,-l}
5 0.869 4.96 0.50 742 0.757 ** 2-fold ( 1-1 0) {-k,-h,-l}
6 0.846 5.52 0.55 719 0.789 ** 2-fold ( 1 1 0) {k,h,-l}
7 0.852 5.44 0.54 1325 1.146 ** 4-fold l ( 0 0 1) {-k,h,l}{k,-h,l}
...
...
Best Solution: space group P 42 21 2
Reindex operator: [k,l,h]
Laue group probability: 0.989
Systematic absence probability: 0.915
Total probability: 0.905
Space group confidence: 0.874
Laue group confidence 0.986
Unit cell: 79.10 79.10 38.30 90.00 90.00 90.00
79.10 to 13.70 - Resolution range used for Laue group search
79.10 to 1.80 - Resolution range in file, used for systematic absence check
Number of batches in file: 3
The data do not appear to be twinned, from the L-test
$$ <!--SUMMARY_END-->
HKLIN spacegroup: P 1 primitive triclinic
$TEXT:Warning:$$ $$
The input crystal system is primitive triclinic
(Cell: 38.30 79.10 79.10 90.00 90.00 90.00)
The crystal system chosen for output is primitive tetragonal
(Cell: 79.10 79.10 38.30 90.00 90.00 90.00)
Based on the P4(2)2(1)2 suggestion, we may try to modify the header of XSCALE.INP to
SPACE_GROUP_NUMBER= 94 UNIT_CELL_CONSTANTS= 79.1 79.1 38.3 90 90 90 OUTPUT_FILE=temp.ahkl SAVE_CORRECTION_IMAGES=FALSE FRIEDEL'S_LAW=TRUE REIDX=0 1 0 0 0 0 1 0 1 0 0 0
where the last line takes care of the shuffling of axes into the order k,l,h, , and obtain
SUBSET OF INTENSITY DATA WITH SIGNAL/NOISE >= -3.0 AS FUNCTION OF RESOLUTION
RESOLUTION NUMBER OF REFLECTIONS COMPLETENESS R-FACTOR R-FACTOR COMPARED I/SIGMA R-meas CC(1/2) Anomal SigAno Nano
LIMIT OBSERVED UNIQUE POSSIBLE OF DATA observed expected Corr
8.03 2978 167 167 100.0% 53.6% 45.8% 2978 5.94 55.1% 99.2* 22 1.190 76
5.68 5488 274 274 100.0% 54.0% 46.1% 5488 6.12 55.4% 97.0* 20 0.915 175
4.64 6976 338 338 100.0% 55.4% 46.1% 6976 6.25 57.0% 99.1* 15 0.983 237
4.01 8069 390 390 100.0% 57.5% 46.3% 8069 6.01 59.0% 93.7* 8 0.991 294
3.59 9191 440 440 100.0% 63.9% 46.7% 9191 5.80 65.5% 89.2* 3 1.071 338
3.28 10239 474 474 100.0% 63.8% 47.0% 10239 5.85 65.4% 89.4* 4 1.119 375
3.03 11037 511 511 100.0% 66.0% 47.5% 11037 5.33 67.6% 91.7* 3 1.068 412
2.84 12014 547 547 100.0% 69.6% 49.1% 12014 4.80 71.2% 82.2* -1 1.092 447
2.68 12698 580 580 100.0% 72.2% 51.0% 12698 4.34 73.9% 83.8* -7 0.969 478
2.54 13360 612 612 100.0% 73.5% 54.1% 13360 3.98 75.3% 73.4* 4 1.025 511
2.42 14299 642 642 100.0% 76.8% 58.2% 14299 3.59 78.6% 57.0* 6 1.016 545
2.32 14827 667 667 100.0% 77.8% 62.3% 14827 3.38 79.6% 70.3* 1 0.924 563
2.23 15588 698 698 100.0% 79.5% 64.6% 15588 3.22 81.3% 64.9* -1 0.914 597
2.15 15888 705 705 100.0% 79.3% 68.0% 15888 3.23 81.1% 52.5* -5 0.882 614
2.07 16867 754 754 100.0% 82.7% 74.7% 16867 2.92 84.6% 50.1* 3 0.920 647
2.01 16847 754 754 100.0% 86.1% 77.3% 16847 2.73 88.1% 47.6* -3 0.839 658
1.95 17842 799 799 100.0% 90.4% 86.7% 17842 2.47 92.4% 49.3* 1 0.822 696
1.89 18095 810 811 99.9% 96.8% 101.2% 18095 2.21 99.1% 44.6* -4 0.773 707
1.84 18633 829 829 100.0% 106.4% 126.3% 18633 1.90 108.9% 39.6* -6 0.730 736
1.80 15510 824 863 95.5% 118.1% 151.4% 15500 1.46 121.2% 32.3* 2 0.688 699
total 256446 11815 11855 99.7% 64.9% 51.6% 256436 3.61 66.5% 97.9* 1 0.910 9805
Analysis with
xscale_isocluster -dim 2 -clu 2 temp.ahkl
yields a iso.pdb which is not at all a single cluster; it is a severely elongated single cloud:
(If the space group were correct, the result of xscale_isocluster should look similar to this:
which is from a lysozyme SSX data collection performed at the SLS; outliers are labelled. In this case, the data are truely tetragonal.)
We must now investigate whether the data have lower than tetragonal symmetry. XSCALEing with
SPACE_GROUP_NUMBER=16 UNIT_CELL_CONSTANTS=38.3 79.1 79.1 90 90 90
gives a new temp.ahkl, with orthorhombic symmetry.
xscale_isocluster -dim 2 -clu 2 temp.ahkl
gives
psi= 0.1692468 nhalo= 0 cluster: 1 center: 2 elements: 51 core: 51 halo: 0 cluster: 2 center: 6 elements: 49 core: 49 halo: 0
and prepares XSCALE.1.INP (and XSCALE.2.INP) for further use (these two files collect the differently, but internally-consistently indexed XDS_ASCII.HKL files).
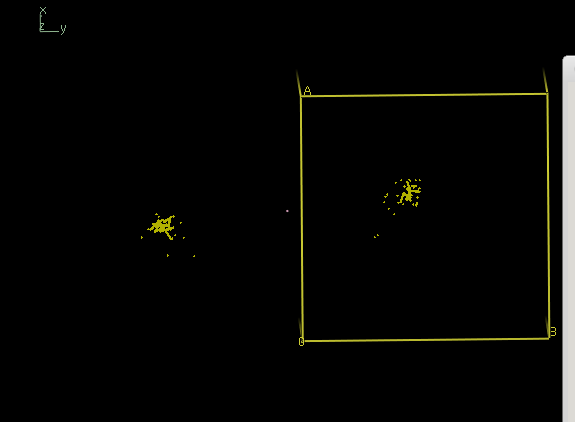
coot iso.pdb
shows
and thus reveals two well separated clouds, corresponding to the two possible indexing modes of the data in space group 19.
Using XSCALE.1.INP with its 51 XDS_ASCII.HKL, and FRIEDEL'S_LAW=TRUE, we get
SUBSET OF INTENSITY DATA WITH SIGNAL/NOISE >= -3.0 AS FUNCTION OF RESOLUTION
RESOLUTION NUMBER OF REFLECTIONS COMPLETENESS R-FACTOR R-FACTOR COMPARED I/SIGMA R-meas CC(1/2) Anomal SigAno Nano
LIMIT OBSERVED UNIQUE POSSIBLE OF DATA observed expected Corr
8.03 1493 297 306 97.1% 11.8% 23.7% 1467 6.04 13.0% 98.2* 52* 0.662 123
5.68 2829 514 521 98.7% 18.9% 24.2% 2796 5.98 20.9% 96.1* 26* 0.778 258
4.64 3576 638 646 98.8% 23.3% 24.2% 3554 6.07 25.7% 93.3* 12 0.829 346
4.01 4140 748 756 98.9% 28.2% 24.5% 4105 5.84 31.0% 89.4* -5 0.818 418
3.59 4735 838 852 98.4% 30.9% 25.0% 4709 5.72 33.9% 86.7* 5 0.983 470
3.28 5268 912 921 99.0% 34.7% 25.8% 5228 5.52 38.0% 85.9* 0 1.005 533
3.03 5664 982 994 98.8% 37.8% 27.4% 5634 4.90 41.4% 82.1* 4 1.031 563
2.84 6114 1065 1068 99.7% 40.4% 31.7% 6082 4.13 44.4% 82.5* 5 0.963 613
2.68 6486 1127 1133 99.5% 44.5% 37.2% 6450 3.54 48.9% 74.8* 1 0.824 644
2.54 6819 1188 1197 99.2% 48.2% 44.6% 6784 3.01 53.0% 70.4* 1 0.816 709
2.42 7278 1249 1259 99.2% 51.9% 54.7% 7249 2.56 56.9% 70.6* 4 0.751 756
2.32 7595 1297 1304 99.5% 55.9% 63.4% 7555 2.26 61.5% 58.5* 4 0.729 809
2.23 7943 1361 1371 99.3% 57.8% 66.4% 7903 2.16 63.3% 63.5* -3 0.687 844
2.15 8093 1375 1385 99.3% 60.1% 75.4% 8054 2.03 65.9% 66.7* 3 0.664 860
2.07 8561 1476 1482 99.6% 64.8% 88.3% 8512 1.76 71.1% 53.0* 7 0.640 914
2.01 8613 1473 1482 99.4% 68.3% 95.8% 8570 1.60 74.9% 60.6* -1 0.628 928
1.95 9048 1566 1571 99.7% 73.1% 112.2% 9004 1.41 80.2% 56.7* -3 0.571 966
1.89 9236 1580 1593 99.2% 82.6% 142.1% 9204 1.19 90.8% 56.3* -5 0.504 1000
1.84 9467 1618 1631 99.2% 92.8% 180.0% 9432 0.96 101.9% 43.2* 4 0.467 1007
1.80 7927 1570 1701 92.3% 104.8% 225.2% 7811 0.70 116.1% 42.6* -5 0.425 785
total 130885 22874 23173 98.7% 38.3% 41.0% 130103 2.77 42.1% 92.0* 3 0.703 13546
At this point, we run
xdscc12 -w XSCALE.1.HKL | grep ^a | sort -nk6
and find that data sets 1 and 17 are wrongly included in the cloud of 51 data sets. Thus they are removed manually from XSCALE.INP.
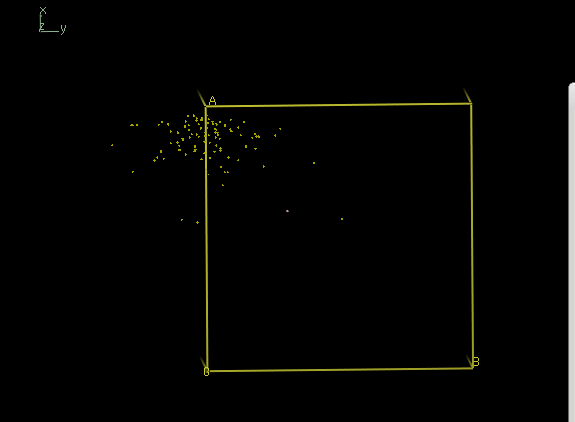
After xscale_isocluster -dim 2 -clu 1 ,
coot iso.pdb
now reveals a single cloud:
We then re-run XSCALE with MERGE=TRUE. The resulting reflection output file XSCALE.1.HKL is then used as REFERENCE_DATA_SET for a second round of integration with XDS.
pointless xdsin XSCALE.1.HKL
gives
Spacegroup TotProb SysAbsProb Reindex Conditions
P 21 21 21 ( 19) 0.896 0.924 h00: h=2n, 0k0: k=2n, 00l: l=2n (zones 1,2,3)
..........
P 2 21 21 ( 18) 0.044 0.045 0k0: k=2n, 00l: l=2n (zones 2,3)
..........
P 21 21 2 ( 18) 0.015 0.015 h00: h=2n, 0k0: k=2n (zones 1,2)
..........
P 21 2 21 ( 18) 0.014 0.014 h00: h=2n, 00l: l=2n (zones 1,3)
---------------------------------------------------------------
Space group confidence (= Sqrt(Score * (Score - NextBestScore))) = 0.87
Laue group confidence (= Sqrt(Score * (Score - NextBestScore))) = 0.97
Selecting space group P 21 21 21 as there is a single space group with the highest score
<!--SUMMARY_BEGIN--> $TEXT:Result: $$ $$
Best Solution: space group P 21 21 21
Reindex operator: [h,k,l]
Laue group probability: 0.970
Systematic absence probability: 0.924
Total probability: 0.896
Space group confidence: 0.874
Laue group confidence 0.966
Unit cell: 38.30 79.10 79.10 90.00 90.00 90.00
79.10 to 2.47 - Resolution range used for Laue group search
79.10 to 1.80 - Resolution range in file, used for systematic absence check
thus we now know the spacegroup.
Round 2: using the REFERENCE_DATA_SET
The processing script integrate.rc is changed a bit, to a) use the REFERENCE_DATA_SET, b) prevent adjustment of variances by CORRECT (this should be done by XSCALE) , c) allow some radiation damage correction in XSCALE:
#!/bin/bash -f
for f in `seq 1 100`;
do
export OUT=wedge0`printf "%03d" $f`
export NAMES="$PWD/Illuin/microfocus/xtal"`printf "%03d" $f`"_1_00\?.img"
rm -rf $OUT
mkdir $OUT
cd $OUT
generate_XDS.INP $NAMES
echo REFERENCE_DATA_SET=../reference.hkl >> XDS.INP
echo MINIMUM_I/SIGMA=50 >>XDS.INP
sed -i s"/SPOT_RANGE=1 1/SPOT_RANGE=1 3/" XDS.INP
sed -i s"/SPACE_GROUP_NUMBER=0/SPACE_GROUP_NUMBER=19/" XDS.INP
sed -i s"/UNIT_CELL_CONSTANTS= 70 80 90/UNIT_CELL_CONSTANTS=38.3 79.1 79.1/" XDS.INP
sed -i s"/TRUSTED_REGION=0.0 1.2/TRUSTED_REGION=0 1/" XDS.INP
sed -i s"/INCLUDE_RESOLUTION_RANGE=50 0/INCLUDE_RESOLUTION_RANGE=99 1.8/" XDS.INP
/usr/local/bin/xds_par
cd ..
done
mkdir xscale
cd xscale
cat >XSCALE.INP <<eof
SPACE_GROUP_NUMBER= 19
UNIT_CELL_CONSTANTS= 38.3 79.1 79.1 90 90 90
OUTPUT_FILE=temp.ahkl
SAVE_CORRECTION_IMAGES=FALSE
eof
find $PWD/../wedge* -name XDS_ASCII.HKL | awk '{print "INPUT_FILE=",$0;print "NBATCH=3 CORRECTIONS=ALL"}' >> XSCALE.INP
and we get as XSCALE.LP :
NOTE: Friedel pairs are treated as different reflections.
SUBSET OF INTENSITY DATA WITH SIGNAL/NOISE >= -3.0 AS FUNCTION OF RESOLUTION
RESOLUTION NUMBER OF REFLECTIONS COMPLETENESS R-FACTOR R-FACTOR COMPARED I/SIGMA R-meas CC(1/2) Anomal SigAno Nano
LIMIT OBSERVED UNIQUE POSSIBLE OF DATA observed expected Corr
8.04 2960 473 476 99.4% 6.2% 5.5% 2955 29.90 6.7% 99.8* 86* 2.824 166
5.68 5486 890 894 99.6% 4.9% 5.9% 5478 27.38 5.3% 99.7* 86* 2.384 363
4.64 6934 1136 1138 99.8% 4.9% 5.8% 6918 27.64 5.4% 99.8* 76* 1.829 480
4.02 8066 1363 1367 99.7% 5.3% 5.9% 8045 26.67 5.9% 99.6* 57* 1.426 590
3.59 9121 1535 1539 99.7% 6.1% 6.3% 9092 25.58 6.7% 99.6* 50* 1.298 666
3.28 10222 1690 1694 99.8% 6.8% 6.8% 10203 24.69 7.5% 99.4* 36* 1.204 751
3.04 10990 1831 1834 99.8% 8.5% 8.0% 10970 21.40 9.3% 99.3* 22* 1.086 827
2.84 12065 1993 1999 99.7% 11.2% 11.1% 12038 17.68 12.2% 99.0* 24* 1.085 894
2.68 12771 2120 2124 99.8% 14.7% 15.1% 12738 14.78 16.1% 98.4* 14* 0.960 952
2.54 13054 2196 2198 99.9% 18.9% 20.2% 13026 12.53 20.8% 97.7* 13* 0.867 995
2.42 14290 2372 2375 99.9% 24.9% 27.1% 14261 10.34 27.3% 96.1* 6 0.813 1083
2.32 14704 2432 2438 99.8% 29.8% 32.5% 14676 9.21 32.6% 95.1* 8 0.843 1115
2.23 15623 2582 2593 99.6% 33.0% 35.0% 15587 8.83 36.1% 93.0* 6 0.831 1180
2.15 15732 2610 2613 99.9% 37.1% 39.2% 15697 8.10 40.6% 91.0* 8 0.818 1203
2.08 16782 2788 2795 99.7% 44.1% 47.0% 16741 7.01 48.3% 88.3* 4 0.797 1276
2.01 16783 2802 2809 99.8% 46.8% 48.7% 16747 6.54 51.2% 89.5* 3 0.807 1293
1.95 18262 3043 3051 99.7% 56.5% 58.0% 18221 5.61 61.9% 85.9* 0 0.803 1402
1.89 17810 2979 2988 99.7% 68.3% 69.8% 17769 4.63 74.8% 80.0* 7 0.864 1374
1.84 18503 3112 3117 99.8% 87.5% 90.3% 18454 3.55 96.0% 69.6* 3 0.838 1435
1.80 16130 2988 3185 93.8% 101.2% 110.5% 15959 2.77 111.7% 62.9* 2 0.798 1276
total 256288 42935 43227 99.3% 13.4% 14.0% 255575 11.63 14.6% 99.6* 21* 0.975 19321
The substructure (locating 4 Se with anom data to 3Å) and structure (198 residues) can now easily be solved with hkl2map:
Further optimization of processing may be possible, but is left as an exercise to the reader.