3CSL: Difference between revisions
(→Peak) |
(→Peak) |
||
| Line 32: | Line 32: | ||
360 frames (0.5° oscillation) at the peak wavelength were collected after the high-remote data. They can be downloaded from [ftp://turn5.biologie.uni-konstanz.de/pub/datasets/3csl-pk.tar here] (1.9 Gb). This peak dataset is somewhat difficult to index; if the results are really bad (e.g. distance refining far away from 370 mm) with the default 180 frames, then just try with 90 or 270 frames. | 360 frames (0.5° oscillation) at the peak wavelength were collected after the high-remote data. They can be downloaded from [ftp://turn5.biologie.uni-konstanz.de/pub/datasets/3csl-pk.tar here] (1.9 Gb). This peak dataset is somewhat difficult to index; if the results are really bad (e.g. distance refining far away from 370 mm) with the default 180 frames, then just try with 90 or 270 frames. | ||
This is an excerpt from [[CORRECT.LP]] : | |||
REFINED PARAMETERS: DISTANCE BEAM ORIENTATION CELL AXIS | |||
USING 166758 INDEXED SPOTS | |||
STANDARD DEVIATION OF SPOT POSITION (PIXELS) 1.80 | |||
STANDARD DEVIATION OF SPINDLE POSITION (DEGREES) 0.61 | |||
CRYSTAL MOSAICITY (DEGREES) 0.557 | |||
DIRECT BEAM COORDINATES (REC. ANGSTROEM) 0.001590 -0.003616 1.021443 | |||
DETECTOR COORDINATES (PIXELS) OF DIRECT BEAM 1536.60 1519.03 | |||
DETECTOR ORIGIN (PIXELS) AT 1528.72 1536.95 | |||
CRYSTAL TO DETECTOR DISTANCE (mm) 370.85 | |||
LAB COORDINATES OF DETECTOR X-AXIS 1.000000 0.000000 0.000000 | |||
LAB COORDINATES OF DETECTOR Y-AXIS 0.000000 1.000000 0.000000 | |||
LAB COORDINATES OF ROTATION AXIS 0.999995 -0.001537 -0.002692 | |||
COORDINATES OF UNIT CELL A-AXIS -41.622 -128.332 80.636 | |||
COORDINATES OF UNIT CELL B-AXIS 45.889 72.744 139.459 | |||
COORDINATES OF UNIT CELL C-AXIS -551.193 220.474 66.370 | |||
REC. CELL PARAMETERS 0.006362 0.006103 0.001674 90.000 90.000 90.000 | |||
UNIT CELL PARAMETERS 157.174 163.848 597.351 90.000 90.000 90.000 | |||
E.S.D. OF CELL PARAMETERS 7.9E-01 8.7E-01 3.0E+00 0.0E+00 0.0E+00 0.0E+00 | |||
SPACE GROUP NUMBER 22 | |||
... | |||
a b ISa | |||
7.764E+00 7.144E-04 13.43 | |||
... | |||
NOTE: Friedel pairs are treated as different reflections. | |||
SUBSET OF INTENSITY DATA WITH SIGNAL/NOISE >= -3.0 AS FUNCTION OF RESOLUTION | |||
RESOLUTION NUMBER OF REFLECTIONS COMPLETENESS R-FACTOR R-FACTOR COMPARED I/SIGMA R-meas Rmrgd-F Anomal SigAno Nano | |||
LIMIT OBSERVED UNIQUE POSSIBLE OF DATA observed expected Corr | |||
8.29 26331 7014 7090 98.9% 5.3% 5.5% 26314 20.22 6.3% 4.2% 71% 1.936 3195 | |||
5.90 48060 12542 12555 99.9% 8.4% 8.4% 48060 13.36 9.8% 8.5% 45% 1.322 5963 | |||
4.83 61534 16074 16144 99.6% 11.0% 10.6% 61534 10.69 12.9% 13.2% 27% 1.051 7728 | |||
4.19 71665 18994 19085 99.5% 12.5% 11.8% 71658 9.61 14.6% 16.0% 18% 0.920 9190 | |||
3.75 77668 21598 21677 99.6% 19.5% 19.3% 77668 6.33 23.0% 27.1% 6% 0.794 10491 | |||
3.42 78594 23767 23865 99.6% 28.3% 29.5% 78582 4.14 34.0% 45.0% 4% 0.735 11548 | |||
3.17 64135 24351 26036 93.5% 42.7% 46.4% 60830 2.18 52.9% 78.5% 2% 0.689 9568 | |||
2.97 40861 20207 27920 72.4% 63.8% 72.3% 35172 1.18 83.3% 118.6% 1% 0.657 6055 | |||
2.80 23238 15074 29715 50.7% 89.5% 104.9% 15359 0.63 122.8% 175.5% 5% 0.646 2502 | |||
total 492086 159621 184087 86.7% 14.8% 15.1% 475177 6.17 17.8% 30.6% 22% 0.901 66240 | |||
NUMBER OF REFLECTIONS IN SELECTED SUBSET OF IMAGES 501334 | |||
NUMBER OF REJECTED MISFITS 8429 | |||
NUMBER OF SYSTEMATIC ABSENT REFLECTIONS 0 | |||
NUMBER OF ACCEPTED OBSERVATIONS 492905 | |||
NUMBER OF UNIQUE ACCEPTED REFLECTIONS 159845 | |||
The verdict is clear: bad ISa, anomalous correlation > 30% only to about 5 A. The reason becomes clear if we load FRAME.cbf into [[XDS-Viewer]], and zoom in: | |||
[[File:3csl-frame.png]] | |||
It is clear that these split reflections provide bad data. Fortunately it seems that in other areas of the detector, the reflections look better. | |||
=== Inflection === | === Inflection === | ||
360 frames (0.5° oscillation) at the inflection wavelength were collected after the peak data. They can be downloaded from [ftp://turn5.biologie.uni-konstanz.de/pub/datasets/3csl-ip.tar here] (1.8 Gb). | 360 frames (0.5° oscillation) at the inflection wavelength were collected after the peak data. They can be downloaded from [ftp://turn5.biologie.uni-konstanz.de/pub/datasets/3csl-ip.tar here] (1.8 Gb). | ||
Revision as of 17:03, 18 March 2011
HasA/R (PDB id 3CSL) is a complex of a 22-stranded beta-barrel outer membrane protein (HAsR, 865 residues), its hemophore (HasA, 206 residues), and heme. The structure and its biological implications are described in "Heme uptake across the outer membrane as revealed by crystal structures of the receptor-hemophore complex" (Krieg, S., Huché, F., Diederichs, K., Izadi-Pruneyre, N., Lecroisey, A., Wandersman, C., Delepelaire, P., Welte, W. (2009), Proc. Nat. Acad. Sci. Vol. 106 pp. 1045-1050.)
3-wl SeMet-MAD data were collected at beamline X06SA of the SLS in November 2006 on a MarCCD detector. HasA/R crystallizes in spacegroup F222; cell parameters are a=157Å, b=163Å, c=596Å. There are 2 complexes per ASU. Data to about 3.0Å could be collected from this crystal, but the anomalous data are useful to about 5Å only. The ordered part of HasR has residues 112-865 and harbours 9 SeMet residues. The ordered part of HasA has 173 residues, one of which is SeMet - but that is mostly disordered.
These MAD data, giving a structure with an average B of 100 Å2, constitute a project that is challenging for humans, and currently too difficult for automatic methods of structure solution and model building. The deposited 3CSL structure was not obtained from these MAD data alone, but the model was actually refined against slightly better (2.7Å) data collected on a native crystal at the ESRF.
XDS data reduction of high-remote, peak and inflection
The script generate_XDS.INP may be used to get a suitable first XDS.INP file for each of the three wavelengths. Unfortunately the beamline software did not put the correct X and Y position of the direct beam into the header. So you will have to find this yourself, using adxv or XDS-viewer. Or just use:
ORGX= 1536 ORGY= 1520
The other thing that you might want to try yourself, or just fill in, is
VALUE_RANGE_FOR_TRUSTED_DETECTOR_PIXELS=8000. 30000. ! often 8000 is ok
instead of the the default (7000. 30000.). This results in a good mask for the beamstop shadow.
It turns out that the spot shapes are actually so irregular that XDS stops after the IDXREF step, with a long warning message. This is because it cannot index (within default error margins) enough reflections (50% is the cutoff). When that occurs, one simply continues with the step after IDXREF:
JOBS= DEFPIX INTEGRATE CORRECT
Other than that, the three MAD wavelengths can be processed once with default parameters, as written into XDS.INP by generate_XDS.INP. This data reduction therefore proceeds in spacegroup P1, but the correct spacegroup (22) is identified by CORRECT.
Optimization: after this first data reduction pass, I use the "post-refined" geometric parameters, and the correct spacegroup (as given in CORRECT.LP, and written to GXPARM.XDS), for a second pass. Thus I need to
mv GXPAM.XDS XPARM.XDS
and modify XDS.INP to read
JOBS= INTEGRATE CORRECT
Afterwards, another xds_par run gives the final intensities. Repeating this optimization sometimes helps.
High-remote
Due to a beamline problem, high-remote data collection stopped after 269 frames of 0.5° (the final frame is already affected). After restart of the beamline, another 100 frames were collected but they later turned out to merge badly with the first 269 frames - a hint that the monochromator was still heating up, or similar. So the latter frames were left out. The 269 frames are here (1.4 Gb).
Peak
360 frames (0.5° oscillation) at the peak wavelength were collected after the high-remote data. They can be downloaded from here (1.9 Gb). This peak dataset is somewhat difficult to index; if the results are really bad (e.g. distance refining far away from 370 mm) with the default 180 frames, then just try with 90 or 270 frames.
This is an excerpt from CORRECT.LP :
REFINED PARAMETERS: DISTANCE BEAM ORIENTATION CELL AXIS
USING 166758 INDEXED SPOTS
STANDARD DEVIATION OF SPOT POSITION (PIXELS) 1.80
STANDARD DEVIATION OF SPINDLE POSITION (DEGREES) 0.61
CRYSTAL MOSAICITY (DEGREES) 0.557
DIRECT BEAM COORDINATES (REC. ANGSTROEM) 0.001590 -0.003616 1.021443
DETECTOR COORDINATES (PIXELS) OF DIRECT BEAM 1536.60 1519.03
DETECTOR ORIGIN (PIXELS) AT 1528.72 1536.95
CRYSTAL TO DETECTOR DISTANCE (mm) 370.85
LAB COORDINATES OF DETECTOR X-AXIS 1.000000 0.000000 0.000000
LAB COORDINATES OF DETECTOR Y-AXIS 0.000000 1.000000 0.000000
LAB COORDINATES OF ROTATION AXIS 0.999995 -0.001537 -0.002692
COORDINATES OF UNIT CELL A-AXIS -41.622 -128.332 80.636
COORDINATES OF UNIT CELL B-AXIS 45.889 72.744 139.459
COORDINATES OF UNIT CELL C-AXIS -551.193 220.474 66.370
REC. CELL PARAMETERS 0.006362 0.006103 0.001674 90.000 90.000 90.000
UNIT CELL PARAMETERS 157.174 163.848 597.351 90.000 90.000 90.000
E.S.D. OF CELL PARAMETERS 7.9E-01 8.7E-01 3.0E+00 0.0E+00 0.0E+00 0.0E+00
SPACE GROUP NUMBER 22
...
a b ISa
7.764E+00 7.144E-04 13.43
...
NOTE: Friedel pairs are treated as different reflections.
SUBSET OF INTENSITY DATA WITH SIGNAL/NOISE >= -3.0 AS FUNCTION OF RESOLUTION
RESOLUTION NUMBER OF REFLECTIONS COMPLETENESS R-FACTOR R-FACTOR COMPARED I/SIGMA R-meas Rmrgd-F Anomal SigAno Nano
LIMIT OBSERVED UNIQUE POSSIBLE OF DATA observed expected Corr
8.29 26331 7014 7090 98.9% 5.3% 5.5% 26314 20.22 6.3% 4.2% 71% 1.936 3195
5.90 48060 12542 12555 99.9% 8.4% 8.4% 48060 13.36 9.8% 8.5% 45% 1.322 5963
4.83 61534 16074 16144 99.6% 11.0% 10.6% 61534 10.69 12.9% 13.2% 27% 1.051 7728
4.19 71665 18994 19085 99.5% 12.5% 11.8% 71658 9.61 14.6% 16.0% 18% 0.920 9190
3.75 77668 21598 21677 99.6% 19.5% 19.3% 77668 6.33 23.0% 27.1% 6% 0.794 10491
3.42 78594 23767 23865 99.6% 28.3% 29.5% 78582 4.14 34.0% 45.0% 4% 0.735 11548
3.17 64135 24351 26036 93.5% 42.7% 46.4% 60830 2.18 52.9% 78.5% 2% 0.689 9568
2.97 40861 20207 27920 72.4% 63.8% 72.3% 35172 1.18 83.3% 118.6% 1% 0.657 6055
2.80 23238 15074 29715 50.7% 89.5% 104.9% 15359 0.63 122.8% 175.5% 5% 0.646 2502
total 492086 159621 184087 86.7% 14.8% 15.1% 475177 6.17 17.8% 30.6% 22% 0.901 66240
NUMBER OF REFLECTIONS IN SELECTED SUBSET OF IMAGES 501334
NUMBER OF REJECTED MISFITS 8429
NUMBER OF SYSTEMATIC ABSENT REFLECTIONS 0
NUMBER OF ACCEPTED OBSERVATIONS 492905
NUMBER OF UNIQUE ACCEPTED REFLECTIONS 159845
The verdict is clear: bad ISa, anomalous correlation > 30% only to about 5 A. The reason becomes clear if we load FRAME.cbf into XDS-Viewer, and zoom in:
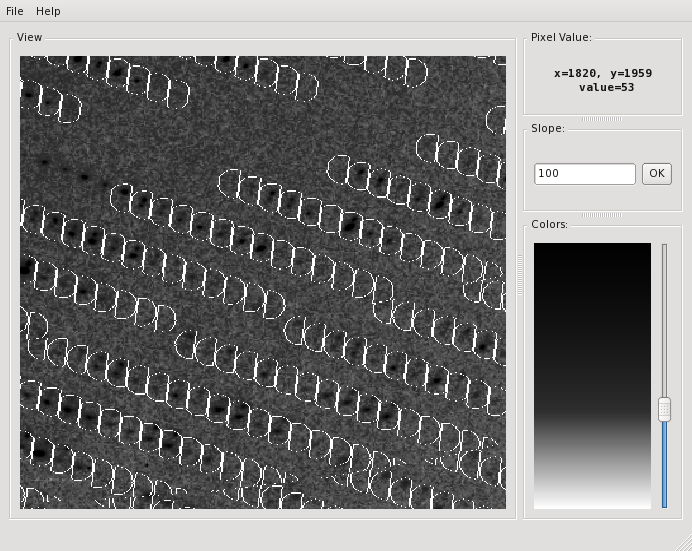 It is clear that these split reflections provide bad data. Fortunately it seems that in other areas of the detector, the reflections look better.
It is clear that these split reflections provide bad data. Fortunately it seems that in other areas of the detector, the reflections look better.
Inflection
360 frames (0.5° oscillation) at the inflection wavelength were collected after the peak data. They can be downloaded from here (1.8 Gb).